Gene Page: DNAH1
Summary ?
| GeneID | 25981 |
| Symbol | DNAH1 |
| Synonyms | DNAHC1|HDHC7|HL-11|HL11|HSRF-1|XLHSRF-1 |
| Description | dynein axonemal heavy chain 1 |
| Reference | MIM:603332|HGNC:HGNC:2940|Ensembl:ENSG00000114841|HPRD:11790|Vega:OTTHUMG00000158378 |
| Gene type | protein-coding |
| Map location | 3p21.1 |
| Pascal p-value | 0.017 |
| Fetal beta | -1.879 |
| eGene | Cerebellar Hemisphere Cerebellum Cortex |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| LK:YES | Genome-wide Association Study | This data set included 99 genes mapped to the 22 regions. The 24 leading SNPs were also included in CV:Ripke_2013 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DNAH1 | chr3 | 52409424 | G | T | NM_015512 | p.2385W>L | missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
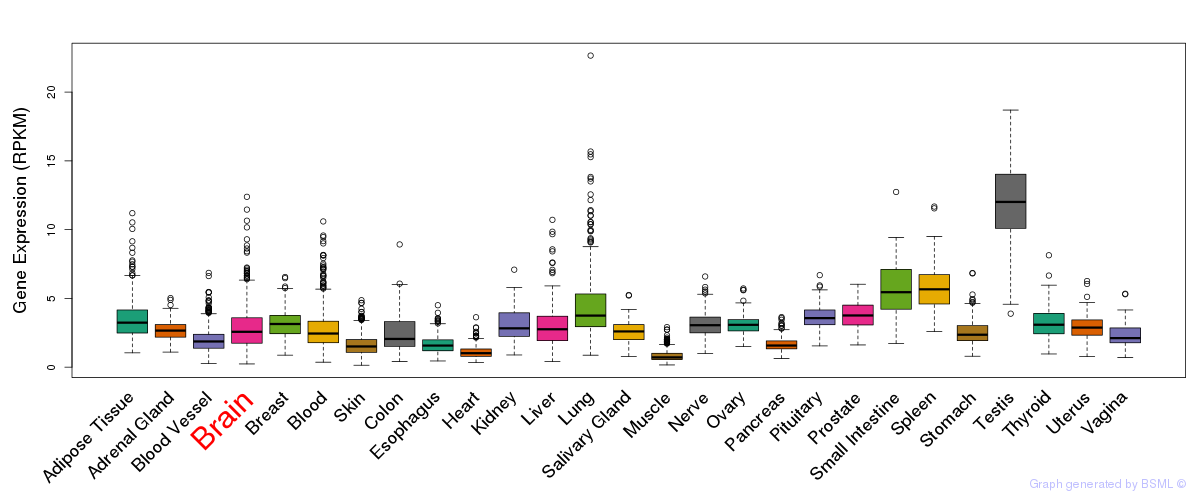
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LDHD | 0.83 | 0.88 |
| GNPTG | 0.82 | 0.83 |
| CLU | 0.80 | 0.84 |
| ALDOC | 0.80 | 0.86 |
| LHPP | 0.80 | 0.83 |
| PTGDS | 0.79 | 0.82 |
| ASPHD1 | 0.79 | 0.81 |
| MVP | 0.78 | 0.82 |
| HLA-DPB1 | 0.77 | 0.82 |
| HDAC11 | 0.77 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZBTB8A | -0.76 | -0.74 |
| KIAA1949 | -0.76 | -0.67 |
| ZNF551 | -0.75 | -0.75 |
| TUBB2B | -0.75 | -0.69 |
| AC004017.1 | -0.75 | -0.74 |
| FAM48A | -0.75 | -0.71 |
| NKIRAS2 | -0.74 | -0.66 |
| SETDB1 | -0.74 | -0.67 |
| ZNF311 | -0.74 | -0.70 |
| MPP3 | -0.74 | -0.67 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| TURASHVILI BREAST NORMAL DUCTAL VS LOBULAR UP | 68 | 40 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATOZOA | 114 | 77 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SUBTIL PROGESTIN TARGETS | 36 | 25 | All SZGR 2.0 genes in this pathway |