Gene Page: DGCR5
Summary ?
| GeneID | 26220 |
| Symbol | DGCR5 |
| Synonyms | LINC00037|NCRNA00037 |
| Description | DiGeorge syndrome critical region gene 5 (non-protein coding) |
| Reference | HGNC:HGNC:16757|Ensembl:ENSG00000237517| |
| Gene type | pseudo |
| Map location | 22q11 |
| Pascal p-value | 0.014 |
| Fetal beta | -1.615 |
| eGene | Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
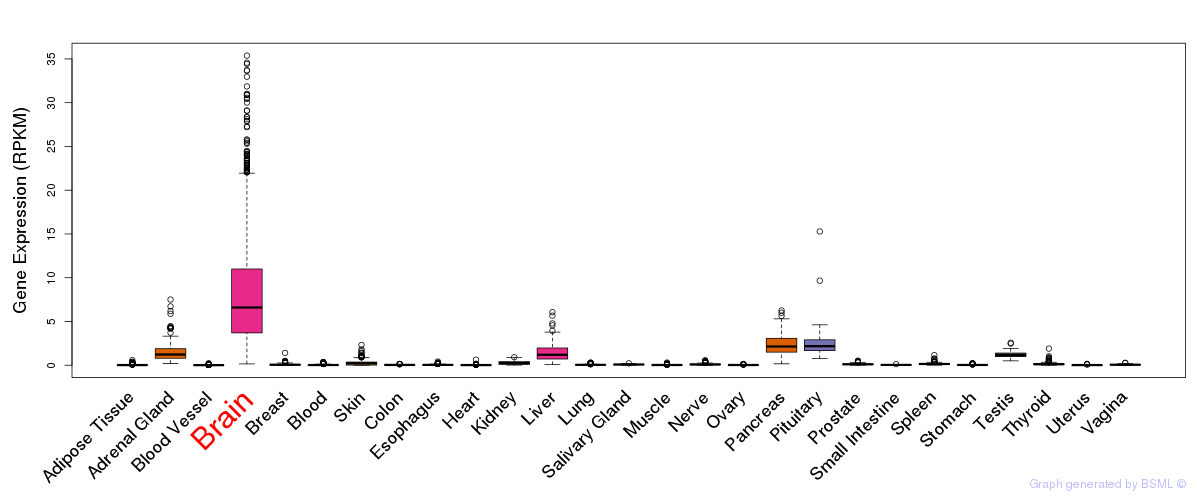
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PAFAH1B3 | 0.94 | 0.83 |
| TUBB2B | 0.93 | 0.88 |
| CCDC101 | 0.93 | 0.77 |
| PPP4C | 0.93 | 0.82 |
| CENPV | 0.92 | 0.82 |
| MARCKSL1 | 0.92 | 0.78 |
| DBN1 | 0.91 | 0.71 |
| SMARCB1 | 0.91 | 0.72 |
| C17orf49 | 0.91 | 0.81 |
| PKN1 | 0.91 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.70 | -0.70 |
| C5orf53 | -0.69 | -0.70 |
| FBXO2 | -0.68 | -0.66 |
| ALDOC | -0.67 | -0.63 |
| CCNI2 | -0.66 | -0.78 |
| CA4 | -0.65 | -0.72 |
| AIFM3 | -0.65 | -0.58 |
| CLU | -0.65 | -0.61 |
| LDHD | -0.65 | -0.63 |
| SPARCL1 | -0.65 | -0.73 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ASTON MAJOR DEPRESSIVE DISORDER UP | 49 | 36 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |