Gene Page: PRICKLE4
Summary ?
| GeneID | 29964 |
| Symbol | PRICKLE4 |
| Synonyms | C6orf49|OBTP|OEBT|TOMM6 |
| Description | prickle planar cell polarity protein 4 |
| Reference | MIM:611389|HGNC:HGNC:16805|Ensembl:ENSG00000124593|HPRD:12884| |
| Gene type | protein-coding |
| Map location | 6p21.31 |
| Pascal p-value | 0.088 |
| Sherlock p-value | 0.232 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.04433 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04163119 | 6 | 41754006 | PRICKLE4 | 5.47E-10 | -0.022 | 8.9E-7 | DMG:Jaffe_2016 |
| cg02125191 | 6 | 41755473 | PRICKLE4 | 1.04E-8 | -0.015 | 4.48E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
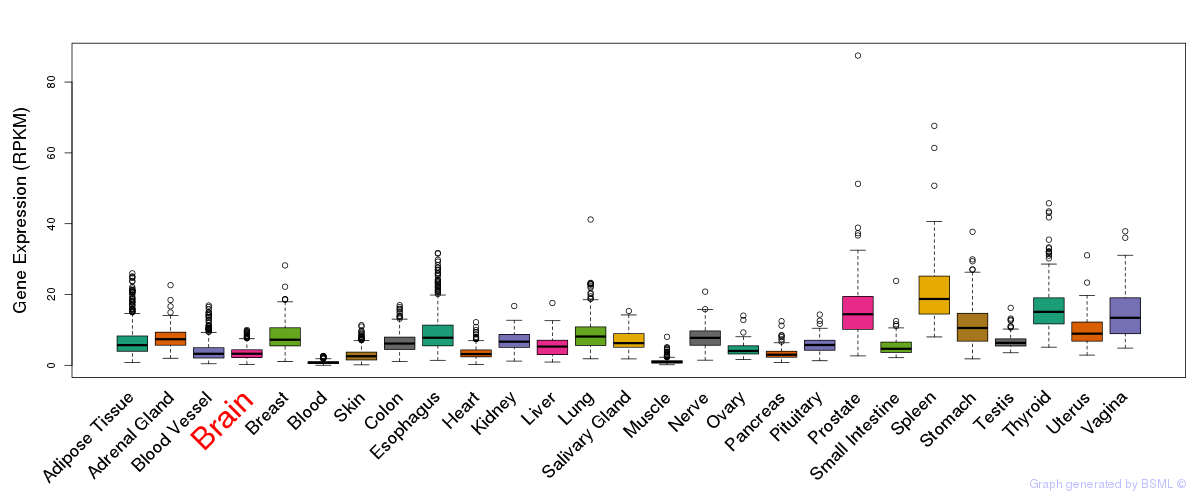
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1219 | 0.91 | 0.96 |
| RAB3GAP2 | 0.91 | 0.94 |
| NUFIP2 | 0.91 | 0.95 |
| BMPR2 | 0.91 | 0.95 |
| TBL1XR1 | 0.91 | 0.94 |
| VCPIP1 | 0.90 | 0.95 |
| APPL1 | 0.90 | 0.94 |
| ATRN | 0.90 | 0.93 |
| SEL1L | 0.90 | 0.94 |
| PRKCI | 0.90 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.64 | -0.70 |
| HIGD1B | -0.60 | -0.69 |
| AF347015.31 | -0.60 | -0.68 |
| MT-CO2 | -0.60 | -0.69 |
| TLCD1 | -0.60 | -0.65 |
| AF347015.21 | -0.59 | -0.68 |
| C19orf36 | -0.59 | -0.64 |
| S100A16 | -0.59 | -0.66 |
| CST3 | -0.59 | -0.66 |
| C1orf54 | -0.58 | -0.72 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| GRADE COLON VS RECTAL CANCER DN | 56 | 36 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-150 | 639 | 645 | 1A | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.