Gene Page: ZC3H6
Summary ?
| GeneID | 376940 |
| Symbol | ZC3H6 |
| Synonyms | ZC3HDC6 |
| Description | zinc finger CCCH-type containing 6 |
| Reference | HGNC:HGNC:24762|Ensembl:ENSG00000188177|HPRD:15698|Vega:OTTHUMG00000153286 |
| Gene type | protein-coding |
| Map location | 2q13 |
| Pascal p-value | 0.146 |
| Sherlock p-value | 0.237 |
| Fetal beta | 1.229 |
| DMG | 1 (# studies) |
| eGene | Meta |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0004 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00755 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ZC3H6 | chr2 | 113080284 | G | C | NM_198581 | p.382R>T | missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14190740 | 2 | 113033328 | ZC3H6 | 4.898E-4 | -0.158 | 0.047 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
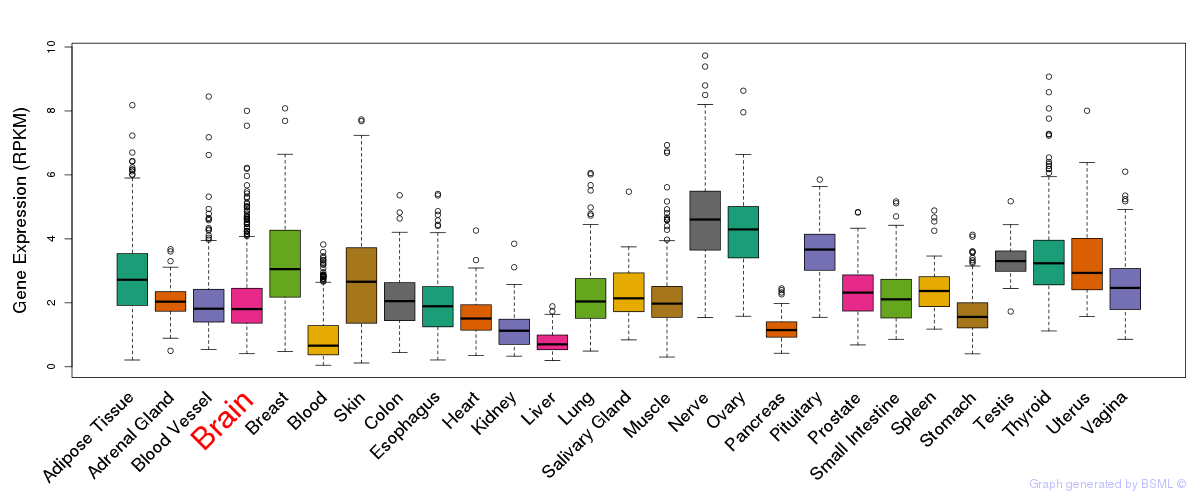
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003676 | nucleic acid binding | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-137 | 357 | 364 | 1A,m8 | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-140 | 128 | 134 | 1A | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-15/16/195/424/497 | 114 | 120 | 1A | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-181 | 574 | 580 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-200bc/429 | 388 | 394 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
| miR-374 | 97 | 103 | 1A | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-410 | 166 | 172 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-433-3p | 828 | 834 | m8 | hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU |
| hsa-miR-433brain | AUCAUGAUGGGCUCCUCGGUGU | ||||
| miR-503 | 114 | 120 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-539 | 2289 | 2295 | 1A | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.