Gene Page: MDK
Summary ?
| GeneID | 4192 |
| Symbol | MDK |
| Synonyms | ARAP|MK|NEGF2 |
| Description | midkine (neurite growth-promoting factor 2) |
| Reference | MIM:162096|HGNC:HGNC:6972|Ensembl:ENSG00000110492|HPRD:01200|Vega:OTTHUMG00000150315 |
| Gene type | protein-coding |
| Map location | 11p11.2 |
| Pascal p-value | 2.765E-10 |
| Sherlock p-value | 0.639 |
| Fetal beta | 0.985 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00078194 | 11 | 46402949 | MDK | 2.84E-5 | -0.2 | 0.018 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17195543 | chr11 | 45509795 | MDK | 4192 | 0.02 | cis | ||
| rs947242 | chr1 | 58042848 | MDK | 4192 | 0.05 | trans | ||
| rs631503 | chr1 | 72327801 | MDK | 4192 | 0.06 | trans | ||
| rs11205098 | chr1 | 152717431 | MDK | 4192 | 6.48E-4 | trans | ||
| rs7518178 | chr1 | 199424428 | MDK | 4192 | 0.18 | trans | ||
| rs3013450 | chr1 | 214552423 | MDK | 4192 | 0.08 | trans | ||
| rs2374323 | chr2 | 41898671 | MDK | 4192 | 5.147E-4 | trans | ||
| rs17486127 | chr2 | 72111878 | MDK | 4192 | 0 | trans | ||
| rs17558168 | chr2 | 72151944 | MDK | 4192 | 0 | trans | ||
| rs7562379 | chr2 | 76693222 | MDK | 4192 | 0.14 | trans | ||
| rs4338920 | chr2 | 141325591 | MDK | 4192 | 0.04 | trans | ||
| rs222809 | chr2 | 146893189 | MDK | 4192 | 0.07 | trans | ||
| rs6726013 | chr2 | 189064570 | MDK | 4192 | 2.414E-4 | trans | ||
| rs6729020 | chr2 | 189064719 | MDK | 4192 | 6.202E-6 | trans | ||
| rs10166301 | chr2 | 189947813 | MDK | 4192 | 0 | trans | ||
| rs4680999 | chr3 | 78613401 | MDK | 4192 | 0.11 | trans | ||
| rs17088743 | chr4 | 58668728 | MDK | 4192 | 0.01 | trans | ||
| rs4425392 | chr4 | 133569611 | MDK | 4192 | 0.17 | trans | ||
| rs35129 | chr5 | 6039853 | MDK | 4192 | 0.09 | trans | ||
| rs447967 | chr5 | 9999056 | MDK | 4192 | 0.14 | trans | ||
| rs6874831 | chr5 | 59155059 | MDK | 4192 | 4.156E-8 | trans | ||
| rs7728346 | chr5 | 59155166 | MDK | 4192 | 8.123E-10 | trans | ||
| rs16890314 | chr5 | 59158770 | MDK | 4192 | 4.597E-7 | trans | ||
| rs10214307 | chr5 | 59159746 | MDK | 4192 | 4.156E-8 | trans | ||
| rs6893381 | chr5 | 59160568 | MDK | 4192 | 9.95E-7 | trans | ||
| rs10052099 | chr5 | 99324715 | MDK | 4192 | 0.05 | trans | ||
| rs10074374 | chr5 | 99324809 | MDK | 4192 | 0.08 | trans | ||
| rs545290 | chr5 | 152858437 | MDK | 4192 | 0.08 | trans | ||
| rs2294670 | chr6 | 625834 | MDK | 4192 | 0.01 | trans | ||
| rs9451867 | chr6 | 92659609 | MDK | 4192 | 0.01 | trans | ||
| rs9640055 | chr7 | 8029515 | MDK | 4192 | 0.05 | trans | ||
| rs17142255 | chr7 | 8075258 | MDK | 4192 | 0.09 | trans | ||
| rs1014201 | chr7 | 25815736 | MDK | 4192 | 0.05 | trans | ||
| rs2939687 | chr8 | 56313877 | MDK | 4192 | 0.11 | trans | ||
| rs10123219 | chr9 | 74156668 | MDK | 4192 | 0.03 | trans | ||
| rs4601388 | chr9 | 94547922 | MDK | 4192 | 1.187E-4 | trans | ||
| rs2006996 | chr9 | 117592637 | MDK | 4192 | 0.16 | trans | ||
| rs2229360 | chr10 | 8116078 | MDK | 4192 | 0.15 | trans | ||
| rs2225536 | chr10 | 110354957 | MDK | 4192 | 0.16 | trans | ||
| rs7933582 | chr11 | 91178991 | MDK | 4192 | 0.14 | trans | ||
| rs7953537 | chr12 | 44693287 | MDK | 4192 | 0.07 | trans | ||
| rs17094401 | chr12 | 44696783 | MDK | 4192 | 0.19 | trans | ||
| rs17094560 | chr12 | 44769767 | MDK | 4192 | 0.07 | trans | ||
| rs8013004 | chr14 | 41098282 | MDK | 4192 | 0.01 | trans | ||
| rs8013037 | chr14 | 41098343 | MDK | 4192 | 0.11 | trans | ||
| rs17105664 | chr14 | 68926192 | MDK | 4192 | 0.18 | trans | ||
| rs1515411 | chr14 | 90614916 | MDK | 4192 | 0.16 | trans | ||
| rs4457950 | chr15 | 35445709 | MDK | 4192 | 0.06 | trans | ||
| rs16969948 | chr15 | 78864785 | MDK | 4192 | 0.14 | trans | ||
| rs4550511 | chr17 | 53340407 | MDK | 4192 | 0.18 | trans | ||
| rs8071763 | chr17 | 67930673 | MDK | 4192 | 3.829E-4 | trans | ||
| rs7244040 | chr18 | 13972023 | MDK | 4192 | 8.92E-8 | trans | ||
| rs17681878 | chr18 | 34767223 | MDK | 4192 | 0.18 | trans | ||
| rs515453 | chr18 | 72483272 | MDK | 4192 | 0.12 | trans | ||
| rs4536719 | chr20 | 24397114 | MDK | 4192 | 8.406E-4 | trans | ||
| rs7887127 | chrX | 38436019 | MDK | 4192 | 1.645E-4 | trans | ||
| rs871948 | chrX | 39342814 | MDK | 4192 | 7.043E-4 | trans |
Section II. Transcriptome annotation
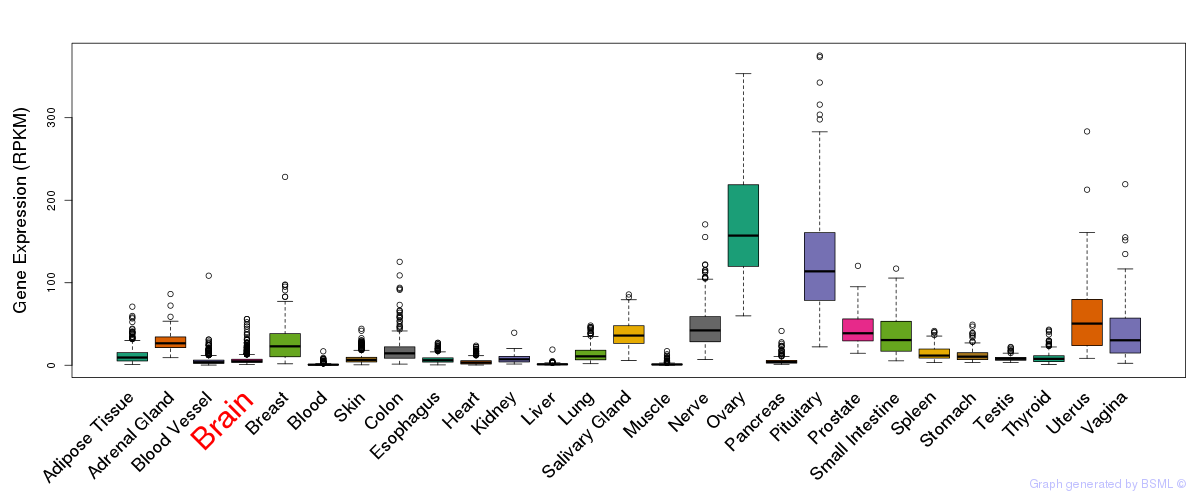
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NEBL | 0.87 | 0.87 |
| EPB41L2 | 0.86 | 0.90 |
| MFAP3L | 0.85 | 0.89 |
| ACSL6 | 0.85 | 0.89 |
| MYLK | 0.85 | 0.82 |
| SYNM | 0.84 | 0.89 |
| SIRPA | 0.84 | 0.91 |
| KIAA0494 | 0.84 | 0.83 |
| DTNA | 0.84 | 0.87 |
| TACC1 | 0.82 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TUBB2B | -0.62 | -0.55 |
| KIAA1949 | -0.62 | -0.49 |
| BCL7C | -0.62 | -0.71 |
| NXT1 | -0.62 | -0.66 |
| RPS8 | -0.61 | -0.75 |
| RPS19P3 | -0.61 | -0.78 |
| RPS13P2 | -0.61 | -0.73 |
| TRAF4 | -0.61 | -0.66 |
| DYNLT1 | -0.61 | -0.74 |
| ALKBH2 | -0.61 | -0.63 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008201 | heparin binding | IDA | 9384573 | |
| GO:0008083 | growth factor activity | IEA | - | |
| GO:0008083 | growth factor activity | NAS | 1639750 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | NAS | neurite (GO term level: 5) | 9384573 |
| GO:0007165 | signal transduction | NAS | 7835084 | |
| GO:0008283 | cell proliferation | IEA | - | |
| GO:0009611 | response to wounding | ISS | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | NAS | 1639750 | |
| GO:0030325 | adrenal gland development | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTG1 | ACT | ACTG | DFNA20 | DFNA26 | actin, gamma 1 | Affinity Capture-MS | BioGRID | 10772929 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD | 9452495 |
| JAK2 | JTK10 | Janus kinase 2 (a protein tyrosine kinase) | - | HPRD | 9452495 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | - | HPRD,BioGRID | 10772929 |
| LRP2 | DBS | gp330 | low density lipoprotein-related protein 2 | - | HPRD | 10772929 |
| NCAM1 | CD56 | MSK39 | NCAM | neural cell adhesion molecule 1 | Affinity Capture-MS | BioGRID | 10772929 |
| NCL | C23 | FLJ45706 | nucleolin | - | HPRD,BioGRID | 12147681 |
| NID1 | NID | nidogen 1 | Affinity Capture-MS | BioGRID | 10772929 |
| NID2 | - | nidogen 2 (osteonidogen) | Affinity Capture-MS | BioGRID | 10772929 |
| PTPRB | DKFZp686E2262 | DKFZp686H15164 | FLJ44133 | HPTP-BETA | HPTPB | MGC142023 | MGC59935 | PTPB | R-PTP-BETA | VEPTP | protein tyrosine phosphatase, receptor type, B | - | HPRD | 10706604 |
| PTPRZ1 | HPTPZ | HPTPzeta | PTP-ZETA | PTP18 | PTPRZ | PTPZ | RPTPB | RPTPbeta | phosphacan | protein tyrosine phosphatase, receptor-type, Z polypeptide 1 | - | HPRD,BioGRID | 10212223 |10706604 |
| RPL18A | - | ribosomal protein L18a | Two-hybrid | BioGRID | 16169070 |
| SDC1 | CD138 | SDC | SYND1 | syndecan | syndecan 1 | - | HPRD | 9089390 |
| SDC3 | N-syndecan | SDCN | SYND3 | syndecan 3 | - | HPRD | 9089390 |
| SDC4 | MGC22217 | SYND4 | syndecan 4 | - | HPRD | 8621465 |
| STAT1 | DKFZp686B04100 | ISGF-3 | STAT91 | signal transducer and activator of transcription 1, 91kDa | - | HPRD | 9452495 |
| TGM1 | ICR2 | KTG | LI | LI1 | TGASE | TGK | transglutaminase 1 (K polypeptide epidermal type I, protein-glutamine-gamma-glutamyltransferase) | - | HPRD | 9384573 |
| TUBA1A | B-ALPHA-1 | FLJ25113 | LIS3 | TUBA3 | tubulin, alpha 1a | Affinity Capture-MS | BioGRID | 10772929 |
| TUBB2B | DKFZp566F223 | FLJ98847 | MGC8685 | TUBB-PARALOG | bA506K6.1 | tubulin, beta 2B | Affinity Capture-MS | BioGRID | 10772929 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID INTEGRIN1 PATHWAY | 66 | 44 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 4 PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE DN | 84 | 53 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK UP | 271 | 175 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 TTD UP | 64 | 39 | All SZGR 2.0 genes in this pathway |
| ROVERSI GLIOMA COPY NUMBER UP | 100 | 75 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| LUI THYROID CANCER CLUSTER 1 | 51 | 33 | All SZGR 2.0 genes in this pathway |
| KORKOLA YOLK SAC TUMOR | 62 | 33 | All SZGR 2.0 genes in this pathway |
| DACOSTA ERCC3 ALLELE XPCS VS TTD UP | 28 | 19 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 UP | 309 | 199 | All SZGR 2.0 genes in this pathway |
| HEIDENBLAD AMPLICON 8Q24 DN | 46 | 28 | All SZGR 2.0 genes in this pathway |
| FURUKAWA DUSP6 TARGETS PCI35 UP | 74 | 32 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS SUBSET | 33 | 20 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB TARGETS | 74 | 41 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR UP | 78 | 56 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 AND CD2 UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS UP | 37 | 27 | All SZGR 2.0 genes in this pathway |
| KANG FLUOROURACIL RESISTANCE UP | 22 | 15 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 24HR DN | 33 | 21 | All SZGR 2.0 genes in this pathway |
| ZHANG ANTIVIRAL RESPONSE TO RIBAVIRIN UP | 30 | 21 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX DN | 88 | 58 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN WITHOUT MGMT 48HR DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 48HR DN | 161 | 105 | All SZGR 2.0 genes in this pathway |
| BACOLOD RESISTANCE TO ALKYLATING AGENTS UP | 26 | 17 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS UP | 24 | 14 | All SZGR 2.0 genes in this pathway |
| KANG CISPLATIN RESISTANCE UP | 19 | 11 | All SZGR 2.0 genes in this pathway |
| SASAI TARGETS OF CXCR6 AND PTCH1 DN | 8 | 6 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| YAMASHITA LIVER CANCER WITH EPCAM UP | 53 | 25 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| CAIRO LIVER DEVELOPMENT UP | 166 | 105 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 12 | 79 | 54 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 UP | 140 | 94 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 90 | 96 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.