Gene Page: ASIP
Summary ?
| GeneID | 434 |
| Symbol | ASIP |
| Synonyms | AGSW|AGTI|AGTIL|ASP|SHEP9 |
| Description | agouti signaling protein |
| Reference | MIM:600201|HGNC:HGNC:745|Ensembl:ENSG00000101440|HPRD:02562|Vega:OTTHUMG00000032289 |
| Gene type | protein-coding |
| Map location | 20q11.2-q12 |
| Pascal p-value | 0.401 |
| Fetal beta | -0.004 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05267394 | 20 | 32856898 | ASIP | 5.75E-4 | 0.959 | 0.049 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
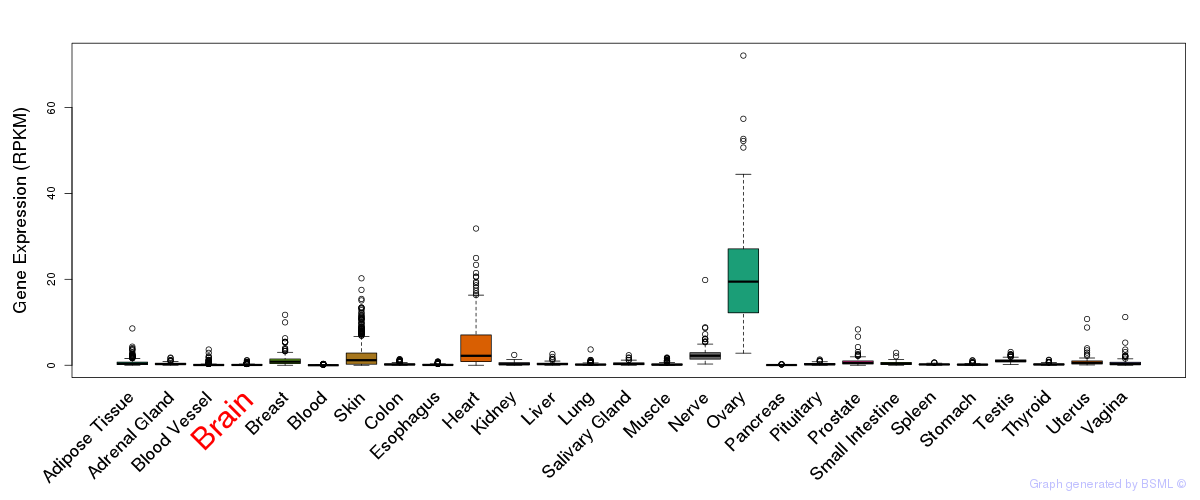
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | TAS | Neurotransmitter (GO term level: 4) | 7757071 |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007267 | cell-cell signaling | TAS | 7757071 | |
| GO:0006091 | generation of precursor metabolites and energy | TAS | 7937887 | |
| GO:0007165 | signal transduction | TAS | 7757071 | |
| GO:0009755 | hormone-mediated signaling | IEA | - | |
| GO:0042438 | melanin biosynthetic process | IEA | - | |
| GO:0043473 | pigmentation | IEA | - | |
| GO:0032438 | melanosome organization | IEA | - | |
| GO:0032402 | melanosome transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | TAS | 7937887 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| PID INSULIN GLUCOSE PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX DN | 80 | 49 | All SZGR 2.0 genes in this pathway |
| LEE CALORIE RESTRICTION NEOCORTEX UP | 83 | 66 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 ICP WITH H3K27ME3 | 74 | 46 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE S | 162 | 86 | All SZGR 2.0 genes in this pathway |