Gene Page: MUSK
Summary ?
| GeneID | 4593 |
| Symbol | MUSK |
| Synonyms | CMS9|FADS |
| Description | muscle, skeletal, receptor tyrosine kinase |
| Reference | MIM:601296|HGNC:HGNC:7525|Ensembl:ENSG00000030304|HPRD:03190|Vega:OTTHUMG00000020485 |
| Gene type | protein-coding |
| Map location | 9q31.3-q32 |
| Pascal p-value | 0.774 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
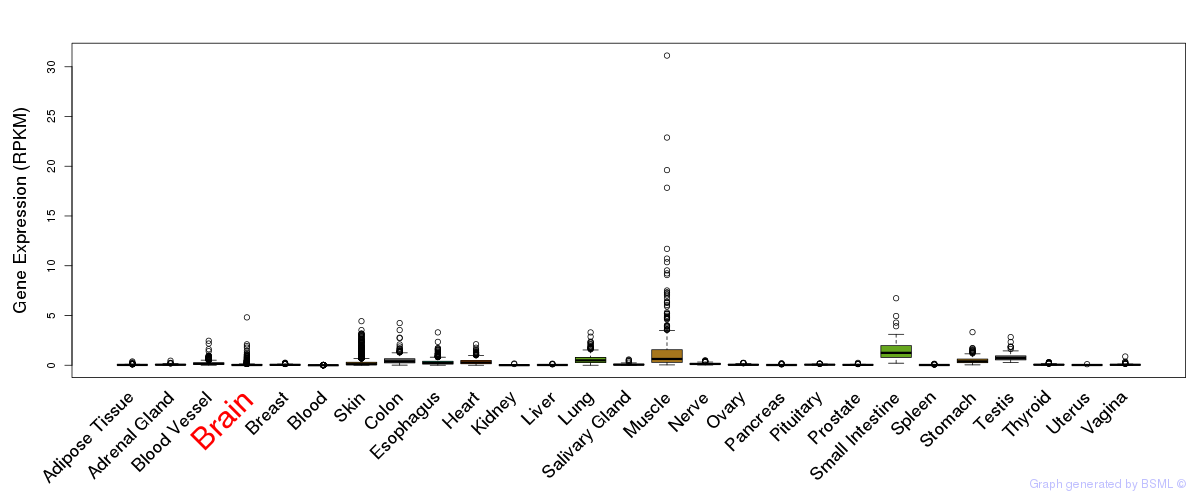
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | TAS | neurite (GO term level: 8) | 7546737 |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008582 | regulation of synaptic growth at neuromuscular junction | IEA | Synap (GO term level: 12) | - |
| GO:0007528 | neuromuscular junction development | IEA | Synap (GO term level: 10) | - |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | TAS | 7546737 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007517 | muscle development | TAS | 7546737 | |
| GO:0007275 | multicellular organismal development | TAS | 7546737 | |
| GO:0043113 | receptor clustering | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0031594 | neuromuscular junction | IEA | neuron, axon, Synap, Neurotransmitter (GO term level: 3) | - |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 7546737 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA AGR PATHWAY | 36 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA ACH PATHWAY | 16 | 13 | All SZGR 2.0 genes in this pathway |
| WIKMAN ASBESTOS LUNG CANCER DN | 28 | 13 | All SZGR 2.0 genes in this pathway |
| DACOSTA LOW DOSE UV RESPONSE VIA ERCC3 XPCS UP | 18 | 10 | All SZGR 2.0 genes in this pathway |
| PETRETTO BLOOD PRESSURE UP | 12 | 8 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE UP | 203 | 130 | All SZGR 2.0 genes in this pathway |
| LABBE WNT3A TARGETS DN | 97 | 53 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 9 11 TRANSLOCATION | 130 | 87 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |