Gene Page: NINJ2
Summary ?
| GeneID | 4815 |
| Symbol | NINJ2 |
| Synonyms | - |
| Description | ninjurin 2 |
| Reference | MIM:607297|HGNC:HGNC:7825|Ensembl:ENSG00000171840|HPRD:09530|Vega:OTTHUMG00000090311 |
| Gene type | protein-coding |
| Map location | 12p13 |
| Pascal p-value | 0.446 |
| Sherlock p-value | 0.066 |
| Fetal beta | -1.873 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4241849 | chr4 | 188964612 | NINJ2 | 4815 | 0.18 | trans | ||
| rs5003961 | chr7 | 78072443 | NINJ2 | 4815 | 0.08 | trans | ||
| rs4515471 | chr7 | 78072585 | NINJ2 | 4815 | 0.08 | trans | ||
| rs10259327 | chr7 | 78074924 | NINJ2 | 4815 | 0.08 | trans | ||
| rs7830259 | chr8 | 14087643 | NINJ2 | 4815 | 8.406E-4 | trans | ||
| rs6482363 | chr10 | 24368607 | NINJ2 | 4815 | 0.06 | trans | ||
| rs1341055 | chr10 | 112607604 | NINJ2 | 4815 | 0.12 | trans | ||
| rs4076909 | chrX | 71329672 | NINJ2 | 4815 | 0.16 | trans |
Section II. Transcriptome annotation
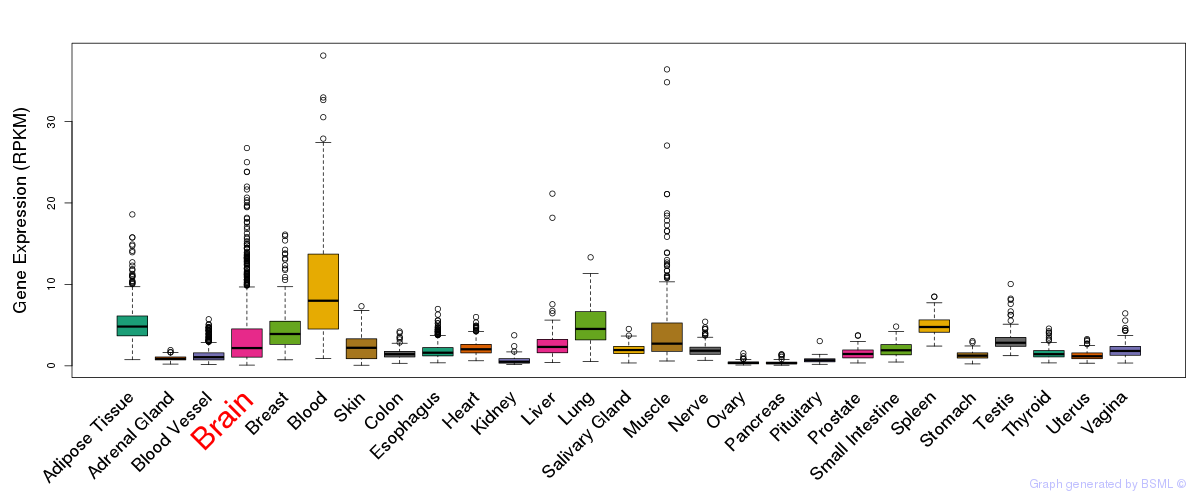
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ENG | 0.78 | 0.78 |
| TIE1 | 0.78 | 0.77 |
| ESAM | 0.77 | 0.75 |
| SCARF1 | 0.76 | 0.80 |
| ROBO4 | 0.76 | 0.78 |
| MMRN2 | 0.76 | 0.76 |
| CDH5 | 0.76 | 0.78 |
| MFSD2 | 0.75 | 0.77 |
| NOTCH4 | 0.73 | 0.75 |
| CXorf36 | 0.73 | 0.75 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MTIF3 | -0.54 | -0.58 |
| RWDD1 | -0.53 | -0.61 |
| CRYZL1 | -0.52 | -0.56 |
| PPP2R3C | -0.50 | -0.55 |
| C8orf59 | -0.50 | -0.59 |
| THOC7 | -0.50 | -0.53 |
| GTPBP10 | -0.49 | -0.51 |
| ENY2 | -0.49 | -0.53 |
| ATP5J | -0.49 | -0.58 |
| CCDC104 | -0.48 | -0.53 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007158 | neuron adhesion | TAS | neuron (GO term level: 5) | 10627596 |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 10627596 |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0042246 | tissue regeneration | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 10627596 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KIM RESPONSE TO TSA AND DECITABINE UP | 129 | 73 | All SZGR 2.0 genes in this pathway |
| GRABARCZYK BCL11B TARGETS DN | 57 | 35 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| ALCALAY AML BY NPM1 LOCALIZATION DN | 184 | 132 | All SZGR 2.0 genes in this pathway |
| HARRIS BRAIN CANCER PROGENITORS | 44 | 23 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 8G | 95 | 62 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS DN | 213 | 127 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |