Gene Page: OSM
Summary ?
| GeneID | 5008 |
| Symbol | OSM |
| Synonyms | - |
| Description | oncostatin M |
| Reference | MIM:165095|HGNC:HGNC:8506|Ensembl:ENSG00000099985|HPRD:01300|Vega:OTTHUMG00000150913 |
| Gene type | protein-coding |
| Map location | 22q12.2 |
| Pascal p-value | 0.017 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs8053315 | chr16 | 83621092 | OSM | 5008 | 0.15 | trans |
Section II. Transcriptome annotation
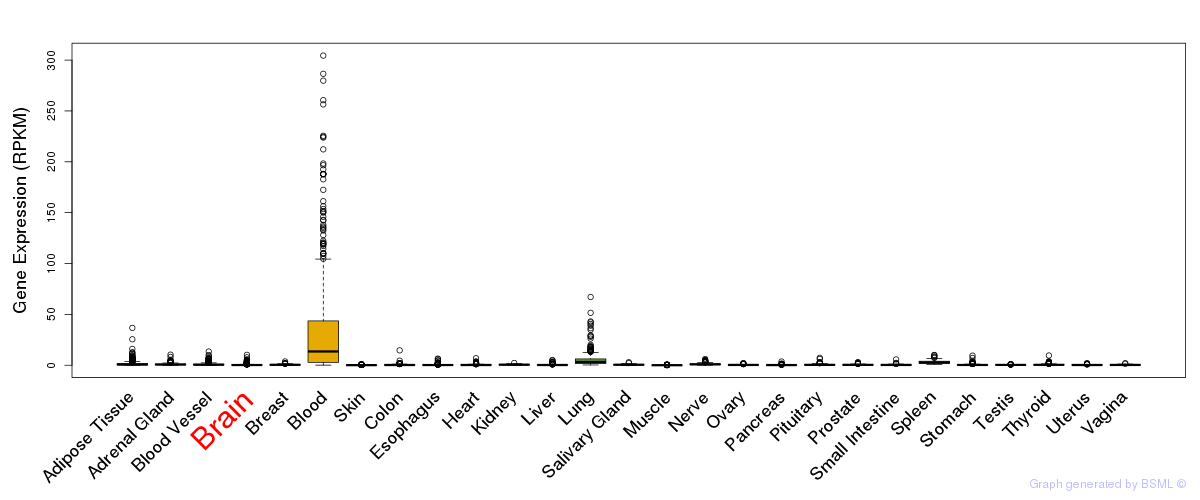
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005147 | oncostatin-M receptor binding | TAS | 9920829 | |
| GO:0005125 | cytokine activity | TAS | 2779549 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001558 | regulation of cell growth | IEA | - | |
| GO:0008285 | negative regulation of cell proliferation | TAS | 1717982 | |
| GO:0009408 | response to heat | IEA | - | |
| GO:0007422 | peripheral nervous system development | IEA | - | |
| GO:0006955 | immune response | IEA | - | |
| GO:0006915 | apoptosis | IEA | - | |
| GO:0007275 | multicellular organismal development | TAS | 1717982 | |
| GO:0033138 | positive regulation of peptidyl-serine phosphorylation | IDA | 7508917 | |
| GO:0042508 | tyrosine phosphorylation of Stat1 protein | IEA | - | |
| GO:0042506 | tyrosine phosphorylation of Stat5 protein | IEA | - | |
| GO:0042503 | tyrosine phosphorylation of Stat3 protein | IEA | - | |
| GO:0048266 | behavioral response to pain | IEA | - | |
| GO:0043410 | positive regulation of MAPKKK cascade | IDA | 7508917 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IDA | 7508917 | |
| GO:0046888 | negative regulation of hormone secretion | IDA | 7867561 | |
| GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | IDA | 7508917 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | TAS | 2779549 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| PID GMCSF PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| PID IL3 PATHWAY | 27 | 19 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP | 485 | 293 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D DN | 31 | 19 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING UP | 108 | 69 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING UP | 115 | 80 | All SZGR 2.0 genes in this pathway |
| VART KSHV INFECTION ANGIOGENIC MARKERS DN | 138 | 92 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 AND H3K27ME3 | 137 | 85 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP | 217 | 131 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-363 | 740 | 746 | 1A | hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA |
| miR-543 | 1006 | 1012 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.