Gene Page: CUZD1
Summary ?
| GeneID | 50624 |
| Symbol | CUZD1 |
| Synonyms | ERG-1|ERG1|ITMAP1|UO-44 |
| Description | CUB and zona pellucida like domains 1 |
| Reference | MIM:616644|HGNC:HGNC:17937|Ensembl:ENSG00000138161|HPRD:06476|Vega:OTTHUMG00000019195 |
| Gene type | protein-coding |
| Map location | 10q26.13 |
| TADA p-value | 0.001 |
| Fetal beta | 0.98 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.03487 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CUZD1 | chr10 | 124593258 | A | T | NM_022034 NR_037912 NR_037915 | p.527Y>* . . | nonsense npcRNA npcRNA | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
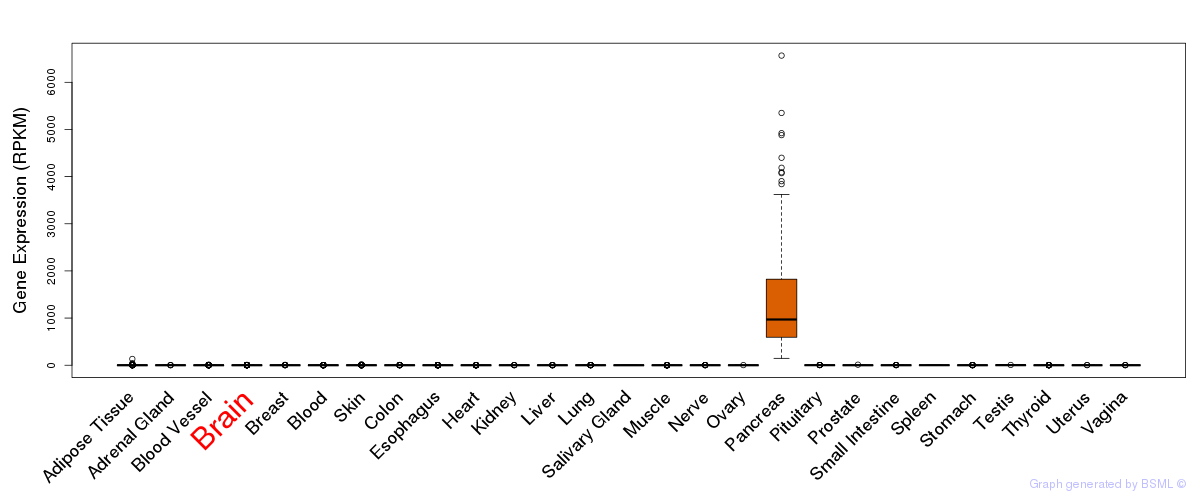
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | IEA | - | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0008283 | cell proliferation | NAS | 15184879 | |
| GO:0006931 | substrate-bound cell migration, cell attachment to substrate | NAS | 15184879 | |
| GO:0051301 | cell division | IEA | - | |
| GO:0032023 | trypsinogen activation | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | NAS | 15184879 | |
| GO:0042589 | zymogen granule membrane | ISS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA CDMAC PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |