Gene Page: PCDHA1
Summary ?
| GeneID | 56147 |
| Symbol | PCDHA1 |
| Synonyms | PCDH-ALPHA1 |
| Description | protocadherin alpha 1 |
| Reference | MIM:606307|HGNC:HGNC:8663|Ensembl:ENSG00000204970|HPRD:07319|Vega:OTTHUMG00000163368 |
| Gene type | protein-coding |
| Map location | 5q31 |
| Pascal p-value | 1.863E-6 |
| eGene | Cerebellar Hemisphere Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGC128 | Genome-wide Association Study | A multi-stage schizophrenia GWAS of up to 36,989 cases and 113,075 controls. Reported by the Schizophrenia Working Group of PGC. 128 independent associations spanning 108 loci | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0032 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 CV:PGC128
CV:PGC128
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|
Section II. Transcriptome annotation
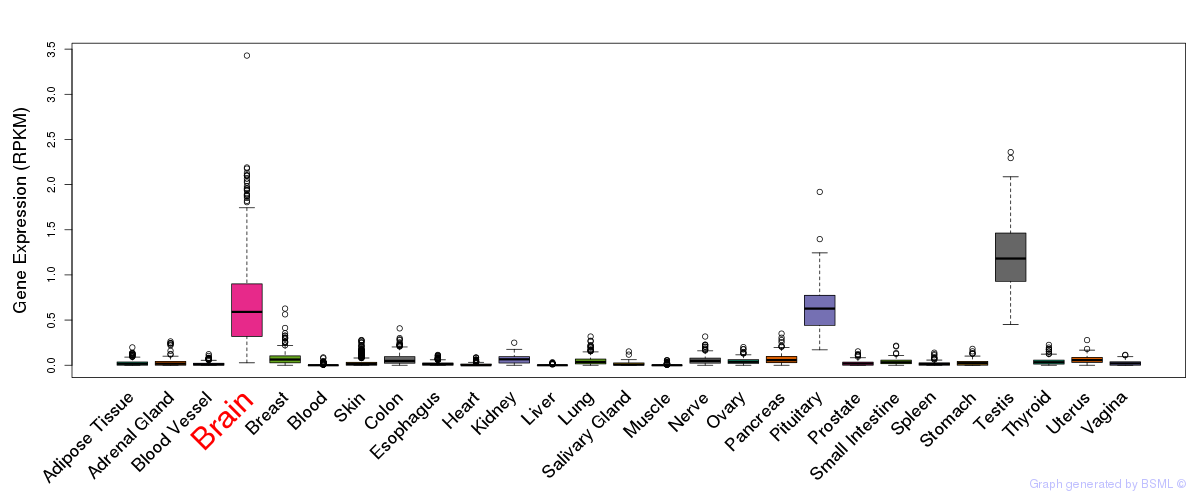
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SHMT1 | 0.76 | 0.65 |
| PKMYT1 | 0.76 | 0.62 |
| ARAF | 0.74 | 0.63 |
| SALL1 | 0.72 | 0.66 |
| PRKD2 | 0.71 | 0.52 |
| HMG20B | 0.71 | 0.57 |
| CHST14 | 0.71 | 0.52 |
| NFKBIL2 | 0.71 | 0.59 |
| RAD54L | 0.71 | 0.48 |
| SHKBP1 | 0.70 | 0.62 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PPEF1 | -0.27 | -0.21 |
| AF347015.31 | -0.27 | -0.17 |
| AF347015.21 | -0.25 | -0.19 |
| AF347015.27 | -0.25 | -0.17 |
| CARD16 | -0.25 | -0.19 |
| MT-CO2 | -0.25 | -0.16 |
| GPR22 | -0.24 | -0.22 |
| CLEC2B | -0.24 | -0.17 |
| AF347015.8 | -0.24 | -0.15 |
| OSTN | -0.24 | -0.20 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 10380929 |
| GO:0007155 | cell adhesion | TAS | 10380929 | |
| GO:0007156 | homophilic cell adhesion | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 10380929 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED | 228 | 119 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 2194 | 2200 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-15/16/195/424/497 | 1015 | 1022 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-153 | 841 | 847 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-17-5p/20/93.mr/106/519.d | 2259 | 2265 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU | ||||
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-181 | 1523 | 1530 | 1A,m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-185 | 204 | 211 | 1A,m8 | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| hsa-miR-185brain | UGGAGAGAAAGGCAGUUC | ||||
| miR-186 | 791 | 797 | m8 | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
| miR-19 | 796 | 803 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-218 | 1791 | 1798 | 1A,m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-221/222 | 60 | 67 | 1A,m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-23 | 2125 | 2131 | 1A | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC | ||||
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-29 | 2355 | 2362 | 1A,m8 | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-320 | 1083 | 1090 | 1A,m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA | ||||
| hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA | ||||
| miR-329 | 2071 | 2078 | 1A,m8 | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-330 | 1833 | 1840 | 1A,m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-362 | 2320 | 2326 | 1A | hsa-miR-362 | AAUCCUUGGAACCUAGGUGUGAGU |
| miR-369-3p | 1057 | 1064 | 1A,m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-448 | 841 | 847 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-495 | 883 | 889 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-503 | 1016 | 1022 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-505 | 1520 | 1526 | 1A | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
| miR-539 | 1257 | 1263 | 1A | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-93.hd/291-3p/294/295/302/372/373/520 | 2258 | 2264 | m8 | hsa-miR-93brain | AAAGUGCUGUUCGUGCAGGUAG |
| hsa-miR-302a | UAAGUGCUUCCAUGUUUUGGUGA | ||||
| hsa-miR-302b | UAAGUGCUUCCAUGUUUUAGUAG | ||||
| hsa-miR-302c | UAAGUGCUUCCAUGUUUCAGUGG | ||||
| hsa-miR-302d | UAAGUGCUUCCAUGUUUGAGUGU | ||||
| hsa-miR-372 | AAAGUGCUGCGACAUUUGAGCGU | ||||
| hsa-miR-373 | GAAGUGCUUCGAUUUUGGGGUGU | ||||
| hsa-miR-520e | AAAGUGCUUCCUUUUUGAGGG | ||||
| hsa-miR-520a | AAAGUGCUUCCCUUUGGACUGU | ||||
| hsa-miR-520b | AAAGUGCUUCCUUUUAGAGGG | ||||
| hsa-miR-520c | AAAGUGCUUCCUUUUAGAGGGUU | ||||
| hsa-miR-520d | AAAGUGCUUCUCUUUGGUGGGUU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.