Gene Page: BARHL1
Summary ?
| GeneID | 56751 |
| Symbol | BARHL1 |
| Synonyms | - |
| Description | BarH like homeobox 1 |
| Reference | MIM:605211|HGNC:HGNC:953|Ensembl:ENSG00000125492|HPRD:05554|Vega:OTTHUMG00000020839 |
| Gene type | protein-coding |
| Map location | 9q34 |
| Pascal p-value | 0.293 |
| Fetal beta | 0.13 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09771468 | 9 | 135462169 | BARHL1 | 2.15E-9 | -0.019 | 1.69E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3765543 | chr9 | 134458323 | BARHL1 | 56751 | 0 | cis | ||
| rs10122864 | chr9 | 134991712 | BARHL1 | 56751 | 0.15 | cis | ||
| rs1009138 | chr9 | 135000607 | BARHL1 | 56751 | 0.09 | cis | ||
| rs17392743 | chr1 | 54764240 | BARHL1 | 56751 | 0.02 | trans | ||
| rs2502827 | chr1 | 176044216 | BARHL1 | 56751 | 0.02 | trans | ||
| rs12405921 | chr1 | 206695730 | BARHL1 | 56751 | 0.15 | trans | ||
| rs17572651 | chr1 | 218943612 | BARHL1 | 56751 | 1.42E-4 | trans | ||
| rs16858054 | chr2 | 171105787 | BARHL1 | 56751 | 0.08 | trans | ||
| rs3845734 | chr2 | 171125572 | BARHL1 | 56751 | 0.01 | trans | ||
| rs8179648 | chr2 | 179377279 | BARHL1 | 56751 | 0.06 | trans | ||
| rs7584986 | chr2 | 184111432 | BARHL1 | 56751 | 6.967E-7 | trans | ||
| rs9810143 | chr3 | 5060209 | BARHL1 | 56751 | 0.19 | trans | ||
| rs7635430 | chr3 | 180152035 | BARHL1 | 56751 | 0.05 | trans | ||
| rs363082 | chr4 | 3132712 | BARHL1 | 56751 | 0.18 | trans | ||
| snp_a-1815771 | 0 | BARHL1 | 56751 | 0.1 | trans | |||
| rs335980 | chr4 | 173329784 | BARHL1 | 56751 | 0.16 | trans | ||
| rs17762315 | chr5 | 76807576 | BARHL1 | 56751 | 0.01 | trans | ||
| rs7729096 | chr5 | 76835927 | BARHL1 | 56751 | 0.01 | trans | ||
| rs10491487 | chr5 | 80323367 | BARHL1 | 56751 | 0.05 | trans | ||
| rs11738579 | chr5 | 95119841 | BARHL1 | 56751 | 0.04 | trans | ||
| rs979348 | chr5 | 97074134 | BARHL1 | 56751 | 0.15 | trans | ||
| rs1368303 | chr5 | 147672388 | BARHL1 | 56751 | 0.09 | trans | ||
| rs13360496 | 0 | BARHL1 | 56751 | 0.07 | trans | |||
| rs16890367 | chr6 | 38078448 | BARHL1 | 56751 | 0.11 | trans | ||
| rs17079498 | chr8 | 3018343 | BARHL1 | 56751 | 0.03 | trans | ||
| rs7021724 | chr9 | 2527899 | BARHL1 | 56751 | 0.13 | trans | ||
| rs11139334 | chr9 | 84209393 | BARHL1 | 56751 | 8.044E-7 | trans | ||
| rs2766987 | chr9 | 113667956 | BARHL1 | 56751 | 0.01 | trans | ||
| rs3765543 | chr9 | 134458323 | BARHL1 | 56751 | 0.14 | trans | ||
| rs2393316 | chr10 | 59333070 | BARHL1 | 56751 | 9.394E-4 | trans | ||
| rs12716680 | chr13 | 42196469 | BARHL1 | 56751 | 0.1 | trans | ||
| rs7996604 | chr13 | 42224412 | BARHL1 | 56751 | 0.04 | trans | ||
| rs7337967 | chr13 | 53519044 | BARHL1 | 56751 | 0.14 | trans | ||
| rs17104720 | chr14 | 77127308 | BARHL1 | 56751 | 0.15 | trans | ||
| rs16955618 | chr15 | 29937543 | BARHL1 | 56751 | 5.892E-31 | trans | ||
| rs2077735 | chr15 | 58479024 | BARHL1 | 56751 | 0.04 | trans | ||
| rs11873184 | chr18 | 1584081 | BARHL1 | 56751 | 0.09 | trans | ||
| rs17761686 | chr18 | 56647634 | BARHL1 | 56751 | 0.07 | trans | ||
| rs7274477 | chr20 | 1676942 | BARHL1 | 56751 | 0.05 | trans | ||
| rs749043 | chr20 | 46995700 | BARHL1 | 56751 | 0.17 | trans | ||
| rs1041786 | chr21 | 22617710 | BARHL1 | 56751 | 2.491E-4 | trans | ||
| rs16990575 | chrX | 32691327 | BARHL1 | 56751 | 0.13 | trans |
Section II. Transcriptome annotation
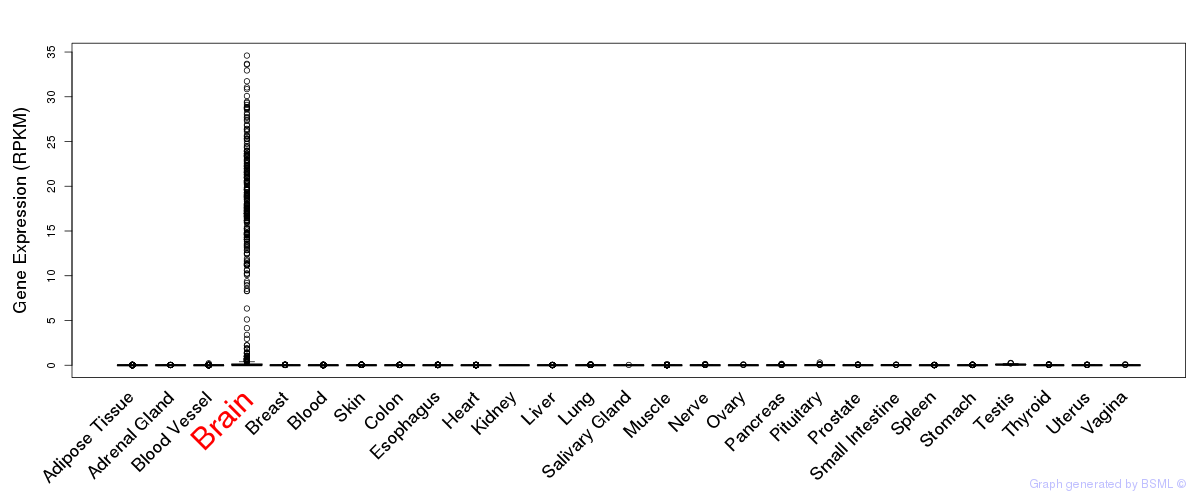
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 17353931 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001764 | neuron migration | IEA | neuron (GO term level: 8) | - |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0007605 | sensory perception of sound | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K27ME3 | 269 | 159 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS HCP WITH H3 UNMETHYLATED | 80 | 50 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-363 | 498 | 504 | 1A | hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA |
| miR-411 | 545 | 551 | m8 | hsa-miR-411 | AACACGGUCCACUAACCCUCAGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.