Gene Page: WRNIP1
Summary ?
| GeneID | 56897 |
| Symbol | WRNIP1 |
| Synonyms | WHIP|bA420G6.2 |
| Description | Werner helicase interacting protein 1 |
| Reference | MIM:608196|HGNC:HGNC:20876|Ensembl:ENSG00000124535|HPRD:10494|Vega:OTTHUMG00000014126 |
| Gene type | protein-coding |
| Map location | 6p25.2 |
| Pascal p-value | 0.372 |
| Sherlock p-value | 0.291 |
| Fetal beta | -0.59 |
| DMG | 1 (# studies) |
| eGene | Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0159 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20466761 | 6 | 2766613 | WRNIP1 | 1.08E-9 | -0.015 | 1.22E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
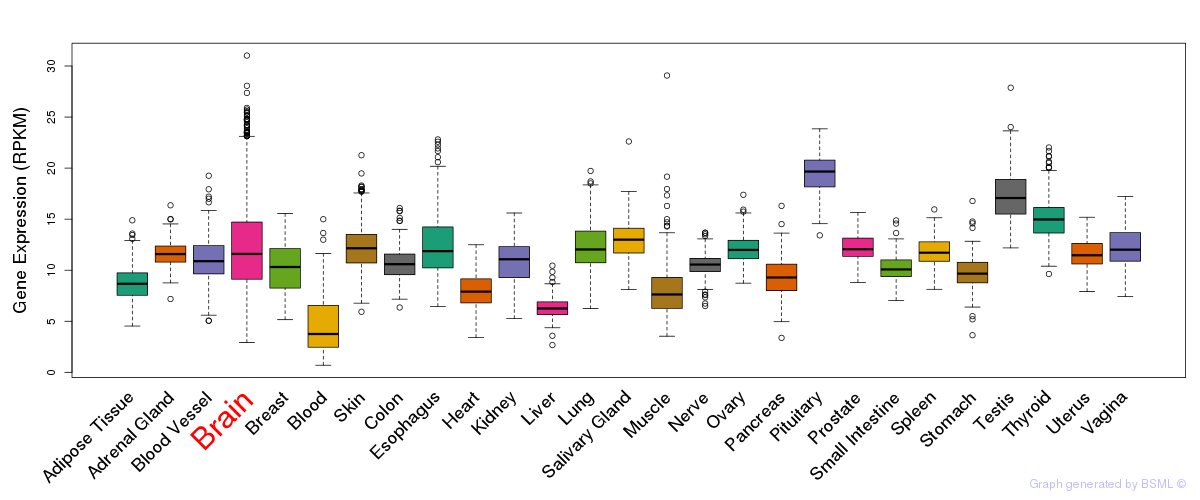
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003677 | DNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 15670210 | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0005524 | ATP binding | IC | 15670210 | |
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0016887 | ATPase activity | IMP | 15670210 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000731 | DNA synthesis during DNA repair | IDA | 15670210 | |
| GO:0030174 | regulation of DNA replication initiation | IDA | 15670210 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | ISS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| VECCHI GASTRIC CANCER EARLY UP | 430 | 232 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPAIR GENES | 230 | 137 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-153 | 423 | 429 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-22 | 229 | 235 | m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-25/32/92/363/367 | 238 | 244 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-320 | 231 | 237 | m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-448 | 422 | 429 | 1A,m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-543 | 250 | 256 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.