Gene Page: PTH2R
Summary ?
| GeneID | 5746 |
| Symbol | PTH2R |
| Synonyms | PTHR2 |
| Description | parathyroid hormone 2 receptor |
| Reference | MIM:601469|HGNC:HGNC:9609|Ensembl:ENSG00000144407|HPRD:03276|Vega:OTTHUMG00000132960 |
| Gene type | protein-coding |
| Map location | 2q33 |
| Pascal p-value | 0.902 |
| Sherlock p-value | 7.698E-4 |
| Fetal beta | -1.088 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00916 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01016 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.009 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25057743 | 2 | 209271505 | PTH2R | 1.94E-8 | -0.014 | 6.81E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
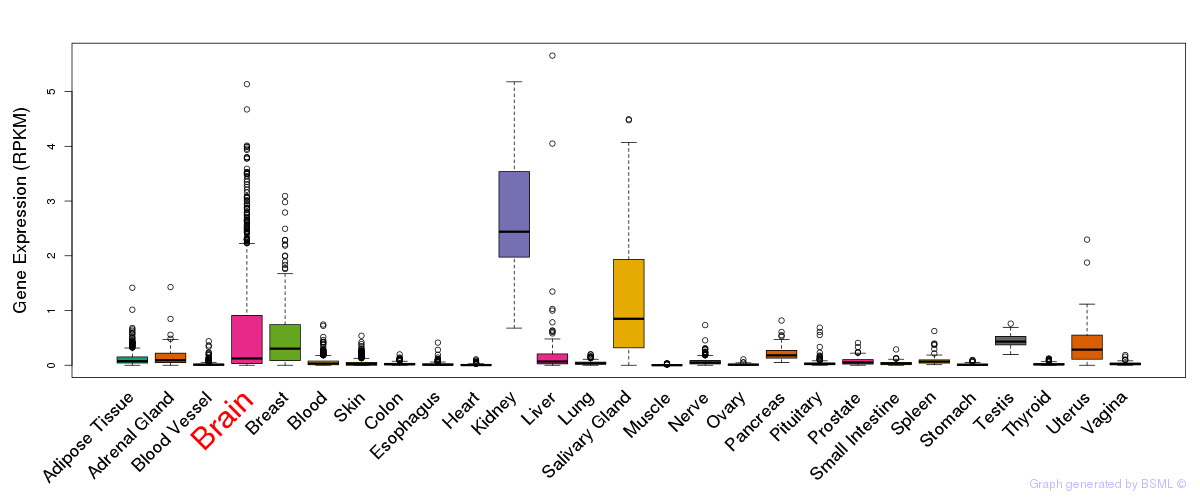
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC005512.1 | 0.91 | 0.21 |
| RGS14 | 0.91 | 0.15 |
| ADORA2A | 0.91 | 0.23 |
| TMEM90A | 0.89 | 0.26 |
| RASD2 | 0.89 | 0.21 |
| WNT6 | 0.88 | 0.23 |
| AP003108.2 | 0.88 | 0.14 |
| DDIT4L | 0.87 | 0.26 |
| ANKRD34B | 0.86 | 0.14 |
| SH3RF2 | 0.86 | 0.31 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MEIS3P2 | -0.24 | -0.14 |
| BLVRA | -0.20 | -0.25 |
| GNAS | -0.18 | -0.05 |
| FXYD6 | -0.18 | -0.19 |
| LRFN2 | -0.18 | -0.16 |
| SH2D3C | -0.17 | -0.08 |
| TUBA1B | -0.17 | -0.17 |
| AC106818.2 | -0.17 | -0.17 |
| RTN4RL2 | -0.17 | -0.14 |
| DYRK3 | -0.17 | -0.18 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004930 | G-protein coupled receptor activity | IEA | - | |
| GO:0004991 | parathyroid hormone receptor activity | TAS | 7797535 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002062 | chondrocyte differentiation | IEA | - | |
| GO:0002076 | osteoblast development | IEA | - | |
| GO:0007200 | G-protein signaling, coupled to IP3 second messenger (phospholipase C activating) | IEA | - | |
| GO:0007189 | G-protein signaling, adenylate cyclase activating pathway | IEA | - | |
| GO:0007165 | signal transduction | IEA | - | |
| GO:0008284 | positive regulation of cell proliferation | IEA | - | |
| GO:0008285 | negative regulation of cell proliferation | IEA | - | |
| GO:0048469 | cell maturation | IEA | - | |
| GO:0006874 | cellular calcium ion homeostasis | IEA | - | |
| GO:0030282 | bone mineralization | IEA | - | |
| GO:0045453 | bone resorption | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 7797535 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | 88 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA S SIGNALLING EVENTS | 121 | 82 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER B | 48 | 22 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G23 UP | 52 | 35 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K4ME3 AND H3K27ME3 | 38 | 34 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 AND H3K27ME3 | 137 | 85 | All SZGR 2.0 genes in this pathway |