Gene Page: NYNRIN
Summary ?
| GeneID | 57523 |
| Symbol | NYNRIN |
| Synonyms | CGIN1|KIAA1305 |
| Description | NYN domain and retroviral integrase containing |
| Reference | HGNC:HGNC:20165|Ensembl:ENSG00000205978|Vega:OTTHUMG00000171314 |
| Gene type | protein-coding |
| Map location | 14q12 |
| Pascal p-value | 0.516 |
| Sherlock p-value | 0.152 |
| Fetal beta | 2.607 |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NYNRIN | chr14 | 24884073 | C | T | NM_025081 | p.1040R>W | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6687849 | chr1 | 175904808 | NYNRIN | 57523 | 0.13 | trans | ||
| rs2481653 | chr1 | 176044120 | NYNRIN | 57523 | 0.17 | trans | ||
| rs2502827 | chr1 | 176044216 | NYNRIN | 57523 | 0.01 | trans | ||
| rs17572651 | chr1 | 218943612 | NYNRIN | 57523 | 0.06 | trans | ||
| rs16829545 | chr2 | 151977407 | NYNRIN | 57523 | 2.59E-19 | trans | ||
| rs16841750 | chr2 | 158288461 | NYNRIN | 57523 | 0.03 | trans | ||
| rs3845734 | chr2 | 171125572 | NYNRIN | 57523 | 5.374E-5 | trans | ||
| rs7584986 | chr2 | 184111432 | NYNRIN | 57523 | 1.899E-13 | trans | ||
| rs6741060 | chr2 | 217169088 | NYNRIN | 57523 | 0.14 | trans | ||
| rs9810143 | chr3 | 5060209 | NYNRIN | 57523 | 0.02 | trans | ||
| rs2183142 | chr4 | 159232695 | NYNRIN | 57523 | 0.06 | trans | ||
| rs170776 | chr4 | 173276735 | NYNRIN | 57523 | 0.11 | trans | ||
| rs1396222 | chr4 | 173279496 | NYNRIN | 57523 | 0.05 | trans | ||
| rs335980 | chr4 | 173329784 | NYNRIN | 57523 | 0.07 | trans | ||
| rs335982 | chr4 | 173330945 | NYNRIN | 57523 | 0.14 | trans | ||
| rs337984 | chr4 | 173411662 | NYNRIN | 57523 | 0.07 | trans | ||
| rs17762315 | chr5 | 76807576 | NYNRIN | 57523 | 0.08 | trans | ||
| rs10491487 | chr5 | 80323367 | NYNRIN | 57523 | 0.03 | trans | ||
| rs9461864 | chr6 | 33481468 | NYNRIN | 57523 | 0.06 | trans | ||
| rs16890367 | chr6 | 38078448 | NYNRIN | 57523 | 0.07 | trans | ||
| rs3118341 | chr9 | 25185518 | NYNRIN | 57523 | 0.02 | trans | ||
| rs9406868 | chr9 | 25223372 | NYNRIN | 57523 | 0.17 | trans | ||
| rs11139334 | chr9 | 84209393 | NYNRIN | 57523 | 0.03 | trans | ||
| rs6480454 | chr10 | 72108174 | NYNRIN | 57523 | 0.15 | trans | ||
| rs12253651 | chr10 | 72109045 | NYNRIN | 57523 | 0.15 | trans | ||
| rs17104720 | chr14 | 77127308 | NYNRIN | 57523 | 0.07 | trans | ||
| rs16955618 | chr15 | 29937543 | NYNRIN | 57523 | 1.787E-32 | trans | ||
| rs11873184 | chr18 | 1584081 | NYNRIN | 57523 | 0.08 | trans | ||
| rs1041786 | chr21 | 22617710 | NYNRIN | 57523 | 0.01 | trans |
Section II. Transcriptome annotation
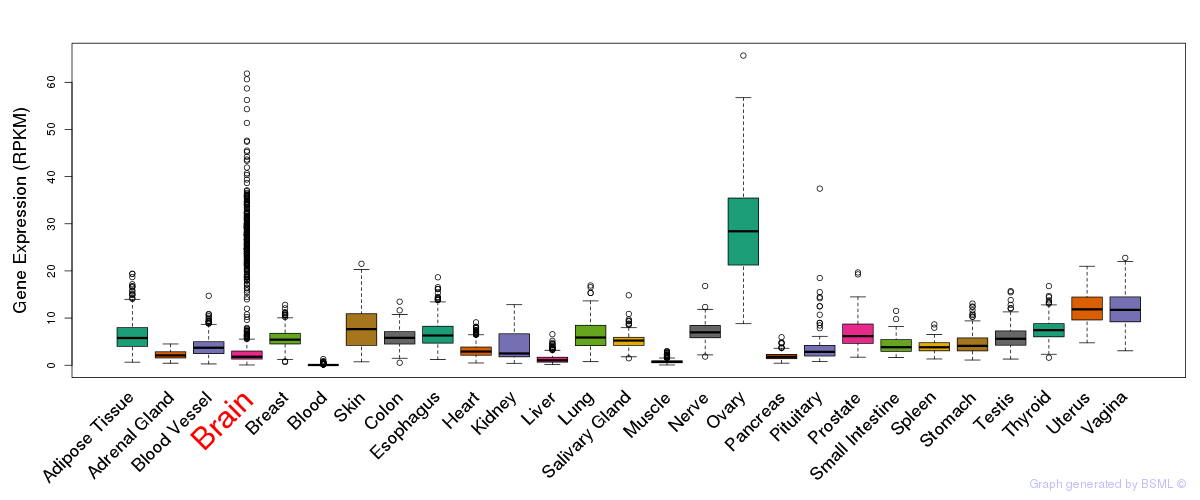
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0015074 | DNA integration | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SOTIRIOU BREAST CANCER GRADE 1 VS 3 DN | 52 | 34 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS DN | 182 | 111 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREY DN | 74 | 44 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION DN | 329 | 219 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR UP | 162 | 116 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE DN | 209 | 137 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 706 | 713 | 1A,m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-124/506 | 103 | 109 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-15/16/195/424/497 | 45 | 52 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-503 | 46 | 52 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.