Gene Page: NEUROD4
Summary ?
| GeneID | 58158 |
| Symbol | NEUROD4 |
| Synonyms | ATH-3|ATH3|Atoh3|MATH-3|MATH3|bHLHa4 |
| Description | neuronal differentiation 4 |
| Reference | MIM:611635|HGNC:HGNC:13802|Ensembl:ENSG00000123307|HPRD:14826|Vega:OTTHUMG00000169826 |
| Gene type | protein-coding |
| Map location | 12q13.2 |
| Pascal p-value | 0.005 |
| Fetal beta | 0.304 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 5 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17532978 | 12 | 55413394 | NEUROD4 | -0.028 | 0.28 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
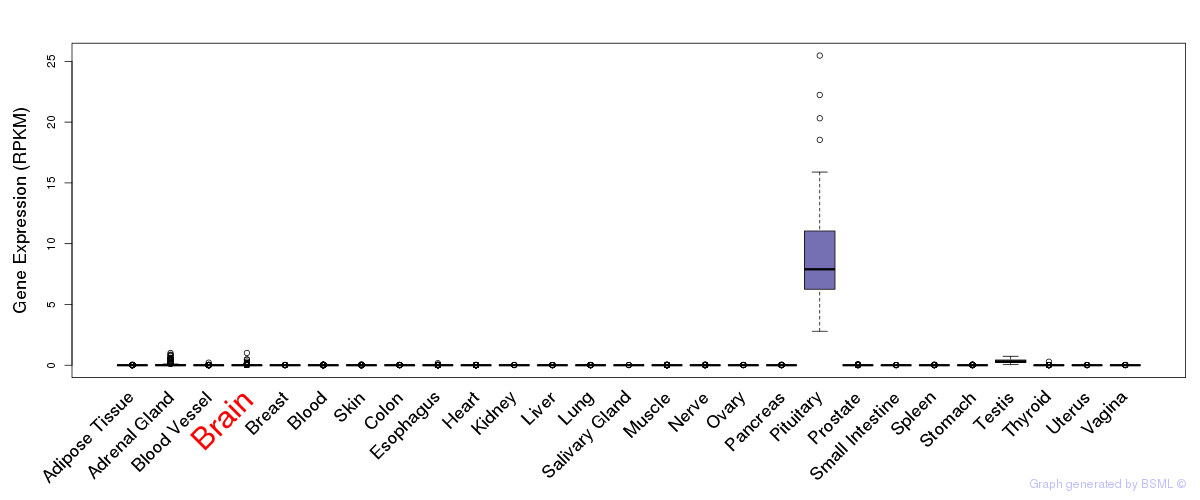
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TMEM126B | 0.84 | 0.81 |
| NDUFB5 | 0.83 | 0.79 |
| TMEM111 | 0.83 | 0.80 |
| CISD1 | 0.83 | 0.82 |
| SDHB | 0.83 | 0.76 |
| SELT | 0.82 | 0.78 |
| MOCS2 | 0.82 | 0.81 |
| NDUFA9 | 0.82 | 0.77 |
| PPA2 | 0.81 | 0.74 |
| AC025647.1 | 0.80 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SH3BP2 | -0.45 | -0.53 |
| SMTN | -0.44 | -0.53 |
| SH2D2A | -0.44 | -0.51 |
| GLIS2 | -0.42 | -0.48 |
| MEGF6 | -0.41 | -0.47 |
| FBN3 | -0.41 | -0.39 |
| TRIOBP | -0.41 | -0.47 |
| AD000671.1 | -0.41 | -0.35 |
| DCHS1 | -0.41 | -0.44 |
| SH2B2 | -0.41 | -0.50 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0030528 | transcription regulator activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007405 | neuroblast proliferation | IEA | neuron (GO term level: 8) | - |
| GO:0001764 | neuron migration | IEA | neuron (GO term level: 8) | - |
| GO:0048666 | neuron development | IEA | neuron (GO term level: 9) | - |
| GO:0010001 | glial cell differentiation | IEA | Glial (GO term level: 8) | - |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0007219 | Notch signaling pathway | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0043010 | camera-type eye development | IEA | - | |
| GO:0045165 | cell fate commitment | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| KIM MYC AMPLIFICATION TARGETS UP | 201 | 127 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT DN | 185 | 111 | All SZGR 2.0 genes in this pathway |
| LEE AGING NEOCORTEX DN | 80 | 49 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |