Gene Page: RIT2
Summary ?
| GeneID | 6014 |
| Symbol | RIT2 |
| Synonyms | RIBA|RIN|ROC2 |
| Description | Ras-like without CAAX 2 |
| Reference | MIM:609592|HGNC:HGNC:10017|Ensembl:ENSG00000152214|HPRD:15253|Vega:OTTHUMG00000132611 |
| Gene type | protein-coding |
| Map location | 18q12.3 |
| Pascal p-value | 0.212 |
| Sherlock p-value | 0.542 |
| Fetal beta | -1.173 |
| eGene | Anterior cingulate cortex BA24 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs438012 | 18 | 39929420 | RIT2 | ENSG00000152214.8 | 3.477E-6 | 0 | 766237 | gtex_brain_ba24 |
| rs371357 | 18 | 39937973 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 757684 | gtex_brain_ba24 |
| rs173199 | 18 | 39948111 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 747546 | gtex_brain_ba24 |
| rs1156542 | 18 | 39953034 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 742623 | gtex_brain_ba24 |
| rs1442648 | 18 | 39954248 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 741409 | gtex_brain_ba24 |
| rs7238485 | 18 | 39955648 | RIT2 | ENSG00000152214.8 | 8.572E-7 | 0 | 740009 | gtex_brain_ba24 |
| rs78272031 | 18 | 39956300 | RIT2 | ENSG00000152214.8 | 2.563E-6 | 0 | 739357 | gtex_brain_ba24 |
| rs1822264 | 18 | 39957067 | RIT2 | ENSG00000152214.8 | 8.572E-7 | 0 | 738590 | gtex_brain_ba24 |
| rs1372762 | 18 | 39962759 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 732898 | gtex_brain_ba24 |
| rs1372763 | 18 | 39962875 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 732782 | gtex_brain_ba24 |
| rs28760399 | 18 | 39964365 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 731292 | gtex_brain_ba24 |
| rs2165714 | 18 | 39965570 | RIT2 | ENSG00000152214.8 | 3.616E-7 | 0 | 730087 | gtex_brain_ba24 |
| rs6507476 | 18 | 39968491 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 727166 | gtex_brain_ba24 |
| rs9304271 | 18 | 39969314 | RIT2 | ENSG00000152214.8 | 3.616E-7 | 0 | 726343 | gtex_brain_ba24 |
| rs398079414 | 18 | 39969448 | RIT2 | ENSG00000152214.8 | 1.281E-6 | 0 | 726209 | gtex_brain_ba24 |
| rs9304272 | 18 | 39970428 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 725229 | gtex_brain_ba24 |
| rs61054461 | 18 | 39970791 | RIT2 | ENSG00000152214.8 | 4.513E-7 | 0 | 724866 | gtex_brain_ba24 |
| rs7233924 | 18 | 39970828 | RIT2 | ENSG00000152214.8 | 8.542E-8 | 0 | 724829 | gtex_brain_ba24 |
| rs346271 | 18 | 39972779 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 722878 | gtex_brain_ba24 |
| rs144588525 | 18 | 39973904 | RIT2 | ENSG00000152214.8 | 3.616E-7 | 0 | 721753 | gtex_brain_ba24 |
| rs77013035 | 18 | 39973905 | RIT2 | ENSG00000152214.8 | 1.304E-6 | 0 | 721752 | gtex_brain_ba24 |
| rs180832 | 18 | 39974236 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 721421 | gtex_brain_ba24 |
| rs189390 | 18 | 39974593 | RIT2 | ENSG00000152214.8 | 8.572E-7 | 0 | 721064 | gtex_brain_ba24 |
| rs346242 | 18 | 39976421 | RIT2 | ENSG00000152214.8 | 2.142E-6 | 0 | 719236 | gtex_brain_ba24 |
| rs6146282 | 18 | 39977236 | RIT2 | ENSG00000152214.8 | 3.392E-7 | 0 | 718421 | gtex_brain_ba24 |
| rs346244 | 18 | 39977576 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 718081 | gtex_brain_ba24 |
| rs346245 | 18 | 39977967 | RIT2 | ENSG00000152214.8 | 8.572E-7 | 0 | 717690 | gtex_brain_ba24 |
| rs346246 | 18 | 39978690 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 716967 | gtex_brain_ba24 |
| rs445617 | 18 | 39979396 | RIT2 | ENSG00000152214.8 | 8.572E-7 | 0 | 716261 | gtex_brain_ba24 |
| rs346247 | 18 | 39981520 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 714137 | gtex_brain_ba24 |
| rs346248 | 18 | 39981791 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 713866 | gtex_brain_ba24 |
| rs346249 | 18 | 39982232 | RIT2 | ENSG00000152214.8 | 3.616E-7 | 0 | 713425 | gtex_brain_ba24 |
| rs346251 | 18 | 39985171 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 710486 | gtex_brain_ba24 |
| rs346252 | 18 | 39986589 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 709068 | gtex_brain_ba24 |
| rs346253 | 18 | 39986664 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 708993 | gtex_brain_ba24 |
| rs346254 | 18 | 39986724 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 708933 | gtex_brain_ba24 |
| rs346255 | 18 | 39987217 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 708440 | gtex_brain_ba24 |
| rs346256 | 18 | 39987515 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 708142 | gtex_brain_ba24 |
| rs346257 | 18 | 39987876 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 707781 | gtex_brain_ba24 |
| rs346258 | 18 | 39988049 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 707608 | gtex_brain_ba24 |
| rs346259 | 18 | 39988232 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 707425 | gtex_brain_ba24 |
| rs346260 | 18 | 39988367 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 707290 | gtex_brain_ba24 |
| rs346261 | 18 | 39988884 | RIT2 | ENSG00000152214.8 | 2.88E-6 | 0 | 706773 | gtex_brain_ba24 |
| rs71174099 | 18 | 39988962 | RIT2 | ENSG00000152214.8 | 2.797E-6 | 0 | 706695 | gtex_brain_ba24 |
| rs346263 | 18 | 39989221 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 706436 | gtex_brain_ba24 |
| rs346264 | 18 | 39989228 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 706429 | gtex_brain_ba24 |
| rs346265 | 18 | 39989431 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 706226 | gtex_brain_ba24 |
| rs189389 | 18 | 39990442 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 705215 | gtex_brain_ba24 |
| rs445364 | 18 | 39991820 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 703837 | gtex_brain_ba24 |
| rs346266 | 18 | 39992444 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 703213 | gtex_brain_ba24 |
| rs346267 | 18 | 39992969 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 702688 | gtex_brain_ba24 |
| rs346268 | 18 | 39993094 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 702563 | gtex_brain_ba24 |
| rs346269 | 18 | 39993389 | RIT2 | ENSG00000152214.8 | 3.616E-7 | 0 | 702268 | gtex_brain_ba24 |
| rs451642 | 18 | 39993631 | RIT2 | ENSG00000152214.8 | 2.701E-6 | 0 | 702026 | gtex_brain_ba24 |
| rs346270 | 18 | 39993663 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 701994 | gtex_brain_ba24 |
| rs180830 | 18 | 39993924 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 701733 | gtex_brain_ba24 |
| rs180831 | 18 | 39994772 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 700885 | gtex_brain_ba24 |
| rs173196 | 18 | 39995744 | RIT2 | ENSG00000152214.8 | 3.616E-7 | 0 | 699913 | gtex_brain_ba24 |
| rs2584798 | 18 | 39996101 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 699556 | gtex_brain_ba24 |
| rs675137 | 18 | 39996712 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 698945 | gtex_brain_ba24 |
| rs417913 | 18 | 39997582 | RIT2 | ENSG00000152214.8 | 1.281E-6 | 0 | 698075 | gtex_brain_ba24 |
| rs410555 | 18 | 39997911 | RIT2 | ENSG00000152214.8 | 2.361E-6 | 0 | 697746 | gtex_brain_ba24 |
| rs437823 | 18 | 39998037 | RIT2 | ENSG00000152214.8 | 1.281E-6 | 0 | 697620 | gtex_brain_ba24 |
| rs397814049 | 18 | 39998578 | RIT2 | ENSG00000152214.8 | 1.241E-6 | 0 | 697079 | gtex_brain_ba24 |
| rs2567734 | 18 | 39999207 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 696450 | gtex_brain_ba24 |
| rs346179 | 18 | 40000484 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 695173 | gtex_brain_ba24 |
| rs173195 | 18 | 40000500 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 695157 | gtex_brain_ba24 |
| rs346178 | 18 | 40000852 | RIT2 | ENSG00000152214.8 | 3.665E-7 | 0 | 694805 | gtex_brain_ba24 |
| rs2567730 | 18 | 40003693 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 691964 | gtex_brain_ba24 |
| rs2567729 | 18 | 40003812 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 691845 | gtex_brain_ba24 |
| rs346181 | 18 | 40008452 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 687205 | gtex_brain_ba24 |
| rs346180 | 18 | 40008628 | RIT2 | ENSG00000152214.8 | 1.281E-6 | 0 | 687029 | gtex_brain_ba24 |
| rs346199 | 18 | 40009943 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 685714 | gtex_brain_ba24 |
| rs346200 | 18 | 40011018 | RIT2 | ENSG00000152214.8 | 1.281E-6 | 0 | 684639 | gtex_brain_ba24 |
| rs346201 | 18 | 40011957 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 683700 | gtex_brain_ba24 |
| rs346202 | 18 | 40012078 | RIT2 | ENSG00000152214.8 | 1.281E-6 | 0 | 683579 | gtex_brain_ba24 |
| rs346204 | 18 | 40013669 | RIT2 | ENSG00000152214.8 | 1.281E-6 | 0 | 681988 | gtex_brain_ba24 |
| rs346206 | 18 | 40013858 | RIT2 | ENSG00000152214.8 | 1.281E-6 | 0 | 681799 | gtex_brain_ba24 |
| rs346208 | 18 | 40014493 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 681164 | gtex_brain_ba24 |
| rs171119 | 18 | 40014887 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 680770 | gtex_brain_ba24 |
| rs184925 | 18 | 40015179 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 680478 | gtex_brain_ba24 |
| rs346209 | 18 | 40016600 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 679057 | gtex_brain_ba24 |
| rs346212 | 18 | 40017917 | RIT2 | ENSG00000152214.8 | 8.572E-7 | 0 | 677740 | gtex_brain_ba24 |
| rs346213 | 18 | 40018447 | RIT2 | ENSG00000152214.8 | 8.572E-7 | 0 | 677210 | gtex_brain_ba24 |
| rs346214 | 18 | 40018754 | RIT2 | ENSG00000152214.8 | 2.733E-6 | 0 | 676903 | gtex_brain_ba24 |
| rs346218 | 18 | 40022060 | RIT2 | ENSG00000152214.8 | 1.038E-6 | 0 | 673597 | gtex_brain_ba24 |
| rs346222 | 18 | 40024675 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 670982 | gtex_brain_ba24 |
| rs346223 | 18 | 40024947 | RIT2 | ENSG00000152214.8 | 8.572E-7 | 0 | 670710 | gtex_brain_ba24 |
| rs5824428 | 18 | 40026912 | RIT2 | ENSG00000152214.8 | 3.6E-6 | 0 | 668745 | gtex_brain_ba24 |
| rs346224 | 18 | 40028311 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 667346 | gtex_brain_ba24 |
| rs346225 | 18 | 40028359 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 667298 | gtex_brain_ba24 |
| rs57552626 | 18 | 40030718 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 664939 | gtex_brain_ba24 |
| rs1431834 | 18 | 40030890 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 664767 | gtex_brain_ba24 |
| rs346182 | 18 | 40031333 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 664324 | gtex_brain_ba24 |
| rs346183 | 18 | 40031488 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 664169 | gtex_brain_ba24 |
| rs237970 | 18 | 40031936 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 663721 | gtex_brain_ba24 |
| rs346184 | 18 | 40032691 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 662966 | gtex_brain_ba24 |
| rs168300 | 18 | 40032866 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 662791 | gtex_brain_ba24 |
| rs346185 | 18 | 40033310 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 662347 | gtex_brain_ba24 |
| rs346186 | 18 | 40033369 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 662288 | gtex_brain_ba24 |
| rs346187 | 18 | 40034530 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 661127 | gtex_brain_ba24 |
| rs346188 | 18 | 40035147 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 660510 | gtex_brain_ba24 |
| rs346189 | 18 | 40035275 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 660382 | gtex_brain_ba24 |
| rs346190 | 18 | 40035793 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 659864 | gtex_brain_ba24 |
| rs346191 | 18 | 40035917 | RIT2 | ENSG00000152214.8 | 2.756E-6 | 0 | 659740 | gtex_brain_ba24 |
| rs346192 | 18 | 40036138 | RIT2 | ENSG00000152214.8 | 2.796E-6 | 0 | 659519 | gtex_brain_ba24 |
| rs375614966 | 18 | 40036171 | RIT2 | ENSG00000152214.8 | 2.774E-6 | 0 | 659486 | gtex_brain_ba24 |
| rs971293 | 18 | 40036223 | RIT2 | ENSG00000152214.8 | 2.828E-6 | 0 | 659434 | gtex_brain_ba24 |
| rs400064 | 18 | 40036313 | RIT2 | ENSG00000152214.8 | 2.931E-6 | 0 | 659344 | gtex_brain_ba24 |
| rs346193 | 18 | 40037280 | RIT2 | ENSG00000152214.8 | 2.151E-6 | 0 | 658377 | gtex_brain_ba24 |
| rs346194 | 18 | 40037616 | RIT2 | ENSG00000152214.8 | 5.213E-9 | 0 | 658041 | gtex_brain_ba24 |
| rs72893569 | 18 | 40092256 | RIT2 | ENSG00000152214.8 | 3.347E-6 | 0 | 603401 | gtex_brain_ba24 |
Section II. Transcriptome annotation
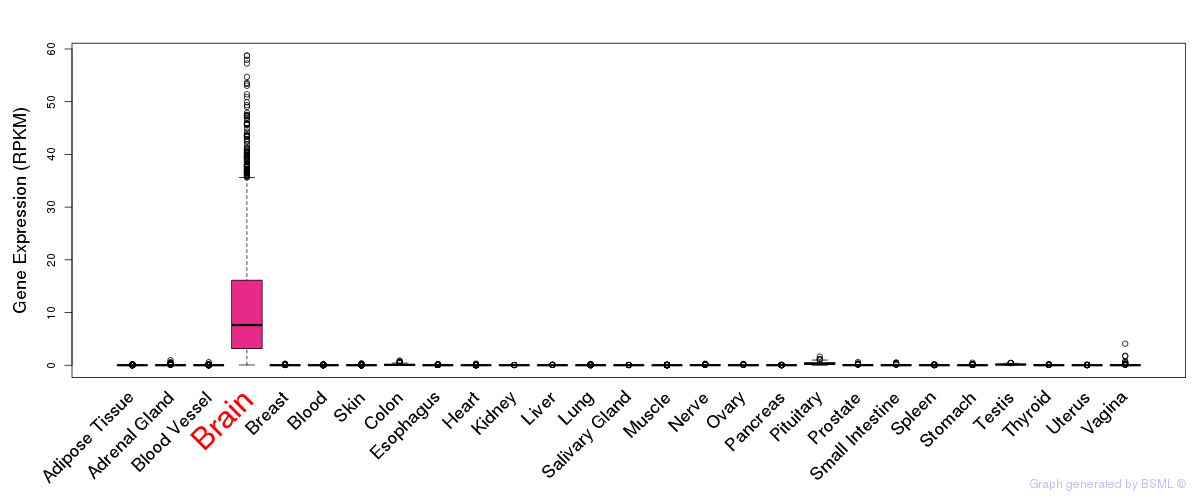
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RNASEN | 0.92 | 0.92 |
| WDR5 | 0.92 | 0.93 |
| SFRS14 | 0.92 | 0.93 |
| PRCC | 0.92 | 0.92 |
| TACC2 | 0.92 | 0.94 |
| UBAP2L | 0.91 | 0.92 |
| LSM14B | 0.91 | 0.93 |
| MSL1 | 0.91 | 0.92 |
| SETD5 | 0.91 | 0.93 |
| TOPBP1 | 0.91 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.86 | -0.89 |
| MT-CO2 | -0.85 | -0.88 |
| AF347015.33 | -0.82 | -0.85 |
| AF347015.27 | -0.82 | -0.86 |
| MT-CYB | -0.82 | -0.85 |
| AF347015.8 | -0.81 | -0.87 |
| HIGD1B | -0.80 | -0.86 |
| AF347015.21 | -0.79 | -0.89 |
| AF347015.2 | -0.79 | -0.86 |
| FXYD1 | -0.79 | -0.84 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003924 | GTPase activity | NAS | 8824319 | |
| GO:0005516 | calmodulin binding | TAS | 8824319 | |
| GO:0005525 | GTP binding | TAS | 8824319 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 8918462 |
| GO:0007165 | signal transduction | TAS | 8918462 | |
| GO:0007264 | small GTPase mediated signal transduction | EXP | 10712923 | |
| GO:0007264 | small GTPase mediated signal transduction | NAS | 8824319 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005886 | plasma membrane | TAS | 8824319 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID TRKR PATHWAY | 62 | 48 | All SZGR 2.0 genes in this pathway |
| PID MAPK TRK PATHWAY | 34 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO ERKS | 36 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO P38 VIA RIT AND RIN | 15 | 14 | All SZGR 2.0 genes in this pathway |
| SILIGAN BOUND BY EWS FLT1 FUSION | 47 | 34 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BANDRES RESPONSE TO CARMUSTIN MGMT 24HR DN | 33 | 21 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL SHORT TERM | 32 | 15 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA RADIATION | 81 | 59 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 UP | 141 | 75 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 250 | 257 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.