Gene Page: SLC5A4
Summary ?
| GeneID | 6527 |
| Symbol | SLC5A4 |
| Synonyms | DJ90G24.4|SAAT1|SGLT3 |
| Description | solute carrier family 5 member 4 |
| Reference | HGNC:HGNC:11039|Ensembl:ENSG00000100191|HPRD:10238|Vega:OTTHUMG00000150007 |
| Gene type | protein-coding |
| Map location | 22q12.3 |
| Pascal p-value | 0.081 |
| Fetal beta | 0.172 |
| eGene | Caudate basal ganglia Cortex Frontal Cortex BA9 Nucleus accumbens basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
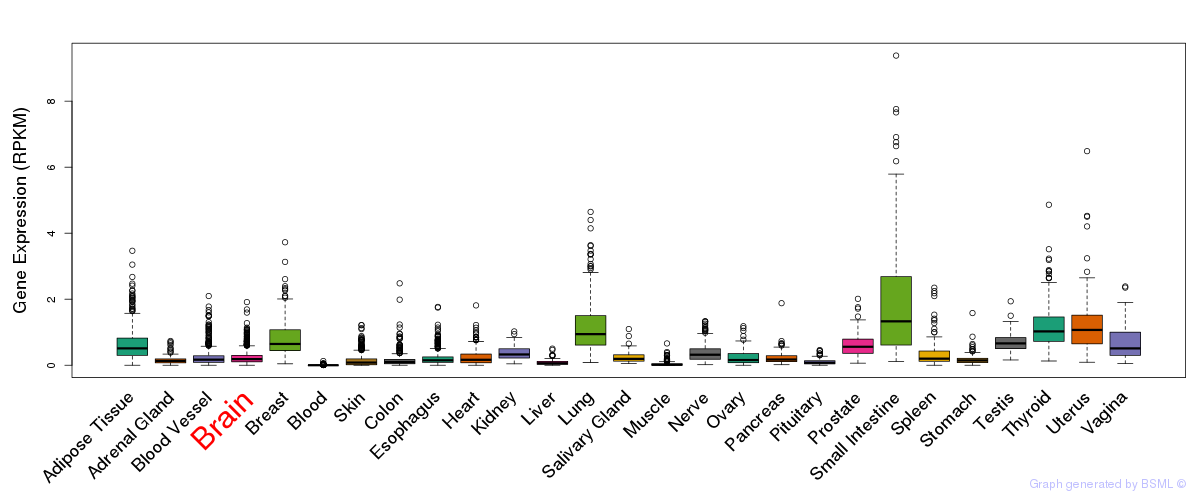
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINA3 | 0.86 | 0.70 |
| CHI3L1 | 0.78 | 0.77 |
| SDS | 0.73 | 0.68 |
| MX1 | 0.72 | 0.68 |
| MT1M | 0.71 | 0.66 |
| MT1X | 0.70 | 0.65 |
| MT1A | 0.67 | 0.59 |
| TMPRSS3 | 0.67 | 0.57 |
| IFITM3 | 0.65 | 0.63 |
| IFIT3 | 0.65 | 0.62 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RTF1 | -0.27 | -0.55 |
| PHF14 | -0.26 | -0.58 |
| PURG | -0.26 | -0.54 |
| AGRN | -0.25 | -0.41 |
| RNF40 | -0.25 | -0.52 |
| ATF5 | -0.25 | -0.49 |
| NOVA2 | -0.25 | -0.52 |
| SAFB | -0.25 | -0.52 |
| AC022098.2 | -0.25 | -0.54 |
| TRIM58 | -0.24 | -0.44 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005351 | sugar:hydrogen symporter activity | IEA | - | |
| GO:0005215 | transporter activity | IEA | - | |
| GO:0015293 | symporter activity | IEA | - | |
| GO:0031402 | sodium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008643 | carbohydrate transport | IEA | - | |
| GO:0006814 | sodium ion transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR DN | 56 | 37 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS UP | 266 | 142 | All SZGR 2.0 genes in this pathway |