Gene Page: SLC17A1
Summary ?
| GeneID | 6568 |
| Symbol | SLC17A1 |
| Synonyms | NAPI-1|NPT-1|NPT1 |
| Description | solute carrier family 17 member 1 |
| Reference | MIM:182308|HGNC:HGNC:10929|Ensembl:ENSG00000124568|HPRD:01661|Vega:OTTHUMG00000016297 |
| Gene type | protein-coding |
| Map location | 6p22.2 |
| Pascal p-value | 5.385E-11 |
| Fetal beta | -0.212 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SLC17A1 | chr6 | 25813095 | C | T | NM_005074 | p.287M>I | missense | Schizophrenia | DNM:Xu_2012 |
Section II. Transcriptome annotation
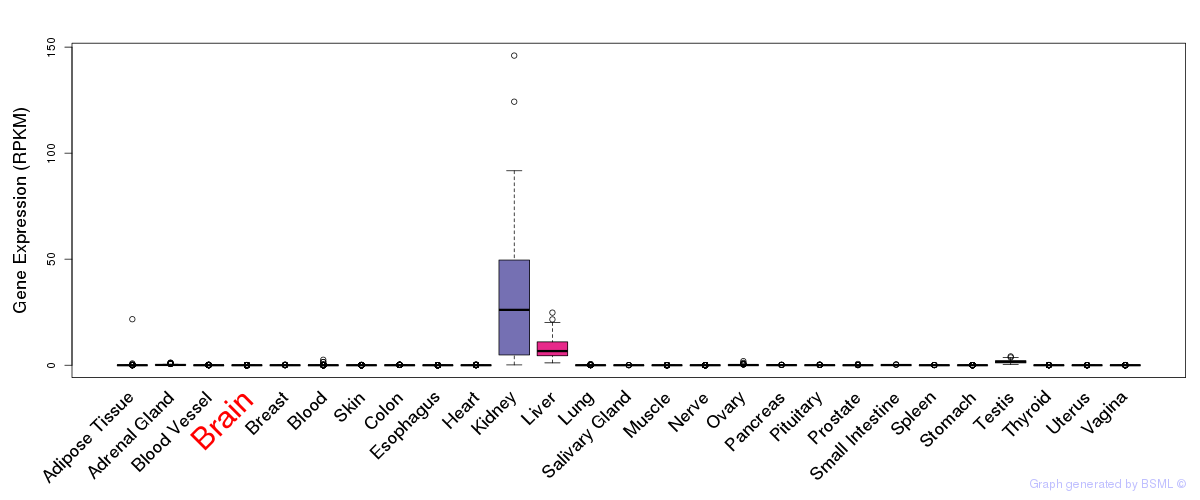
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CHST14 | 0.92 | 0.82 |
| PTBP1 | 0.92 | 0.78 |
| TGIF2 | 0.92 | 0.78 |
| TP53 | 0.91 | 0.76 |
| EPHB4 | 0.90 | 0.80 |
| ERBB2 | 0.90 | 0.87 |
| TEAD2 | 0.89 | 0.66 |
| FZD2 | 0.89 | 0.77 |
| PRKD2 | 0.89 | 0.69 |
| CENPM | 0.87 | 0.51 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SERPINI1 | -0.40 | -0.53 |
| CHN1 | -0.39 | -0.47 |
| ADAP1 | -0.38 | -0.39 |
| EPHX4 | -0.38 | -0.54 |
| CCNI2 | -0.38 | -0.44 |
| OSBPL1A | -0.38 | -0.44 |
| OSTF1 | -0.38 | -0.47 |
| CKMT1A | -0.37 | -0.45 |
| HTATIP2 | -0.37 | -0.42 |
| C5orf53 | -0.37 | -0.38 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005215 | transporter activity | IEA | - | |
| GO:0015293 | symporter activity | IEA | - | |
| GO:0015321 | sodium-dependent phosphate transmembrane transporter activity | TAS | 8867793 | |
| GO:0031402 | sodium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006817 | phosphate transport | TAS | 8867793 | |
| GO:0006814 | sodium ion transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005624 | membrane fraction | TAS | 8288239 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 8288239 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME SLC MEDIATED TRANSMEMBRANE TRANSPORT | 241 | 157 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | 94 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | 49 | 36 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| SU KIDNEY | 15 | 8 | All SZGR 2.0 genes in this pathway |
| ZHENG IL22 SIGNALING DN | 42 | 26 | All SZGR 2.0 genes in this pathway |
| PIONTEK PKD1 TARGETS UP | 38 | 24 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 UP | 176 | 110 | All SZGR 2.0 genes in this pathway |