Gene Page: STX4
Summary ?
| GeneID | 6810 |
| Symbol | STX4 |
| Synonyms | STX4A|p35-2 |
| Description | syntaxin 4 |
| Reference | MIM:186591|HGNC:HGNC:11439|Ensembl:ENSG00000103496|HPRD:01722|Vega:OTTHUMG00000132404 |
| Gene type | protein-coding |
| Map location | 16p11.2 |
| Pascal p-value | 0.31 |
| Sherlock p-value | 0.009 |
| Fetal beta | -0.392 |
| eGene | Cerebellum Frontal Cortex BA9 Nucleus accumbens basal ganglia Meta |
| Support | EXOCYTOSIS |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.015 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
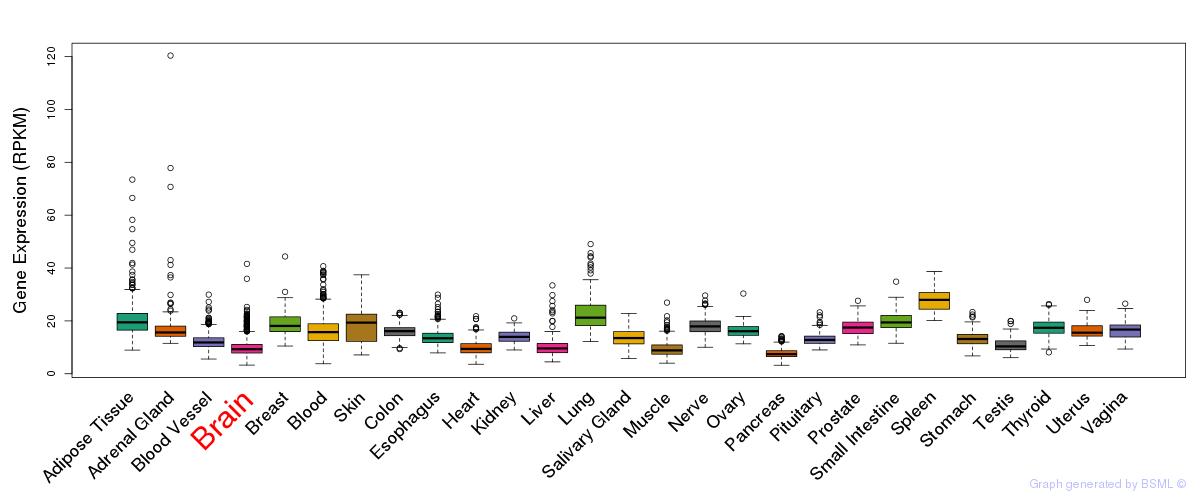
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DOCK2 | 0.84 | 0.76 |
| CD68 | 0.84 | 0.73 |
| ITGB2 | 0.83 | 0.77 |
| NCKAP1L | 0.83 | 0.75 |
| HCK | 0.82 | 0.69 |
| SIGLEC10 | 0.79 | 0.71 |
| LCP1 | 0.79 | 0.71 |
| CX3CR1 | 0.78 | 0.78 |
| CSF1R | 0.77 | 0.79 |
| SASH3 | 0.77 | 0.61 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.27 | -0.33 |
| FXYD1 | -0.27 | -0.30 |
| CXCL14 | -0.27 | -0.26 |
| MT1G | -0.26 | -0.30 |
| S100A13 | -0.26 | -0.26 |
| MYL3 | -0.25 | -0.27 |
| AC098691.2 | -0.25 | -0.31 |
| AF347015.18 | -0.25 | -0.30 |
| CSAG1 | -0.25 | -0.29 |
| AF347015.21 | -0.25 | -0.29 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005484 | SNAP receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006836 | neurotransmitter transport | IEA | neuron, Neurotransmitter (GO term level: 5) | - |
| GO:0006886 | intracellular protein transport | IEA | - | |
| GO:0016192 | vesicle-mediated transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10856305 | |
| GO:0005773 | vacuole | TAS | 16339081 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 18029348 | |
| GO:0016323 | basolateral plasma membrane | IDA | 16339081 | |
| GO:0031012 | extracellular matrix | IDA | 18029348 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| NAPA | SNAPA | N-ethylmaleimide-sensitive factor attachment protein, alpha | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| NSF | SKD2 | N-ethylmaleimide-sensitive factor | - | HPRD | 8973549 |
| RAB11A | MGC1490 | YL8 | RAB11A, member RAS oncogene family | - | HPRD | 12435603 |
| RAB4A | HRES-1/RAB4 | RAB4 | RAB4A, member RAS oncogene family | - | HPRD,BioGRID | 11063739 |
| RAB5A | RAB5 | RAB5A, member RAS oncogene family | - | HPRD,BioGRID | 11884531 |
| SEPT5 | CDCREL | CDCREL-1 | CDCREL1 | H5 | PNUTL1 | septin 5 | - | HPRD,BioGRID | 11880646 |
| SNAP23 | HsT17016 | SNAP23A | SNAP23B | synaptosomal-associated protein, 23kDa | - | HPRD,BioGRID | 9168999 |
| SNAP25 | FLJ23079 | RIC-4 | RIC4 | SEC9 | SNAP | SNAP-25 | bA416N4.2 | dJ1068F16.2 | synaptosomal-associated protein, 25kDa | - | HPRD | 8760387 |
| SNAP25 | FLJ23079 | RIC-4 | RIC4 | SEC9 | SNAP | SNAP-25 | bA416N4.2 | dJ1068F16.2 | synaptosomal-associated protein, 25kDa | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 7768895 |8663154 |9852078 |10194441 |
| SNAP29 | CEDNIK | FLJ21051 | SNAP-29 | synaptosomal-associated protein, 29kDa | Reconstituted Complex | BioGRID | 9852078 |
| STXBP1 | EIEE4 | MUNC18-1 | UNC18 | hUNC18 | rbSec1 | syntaxin binding protein 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 7768895 |12773094 |
| STXBP2 | Hunc18b | MUNC18-2 | UNC18-2 | UNC18B | pp10122 | syntaxin binding protein 2 | Reconstituted Complex | BioGRID | 7768895 |
| STXBP3 | MUNC18-3 | MUNC18C | PSP | UNC-18C | syntaxin binding protein 3 | - | HPRD | 10194441 |
| STXBP3 | MUNC18-3 | MUNC18C | PSP | UNC-18C | syntaxin binding protein 3 | Affinity Capture-Western Two-hybrid | BioGRID | 12773094 |12832401 |
| STXBP4 | FLJ16496 | MGC149829 | MGC50337 | Synip | syntaxin binding protein 4 | - | HPRD | 10394363 |12855681 |
| STXBP5 | FLJ30922 | LGL3 | LLGL3 | MGC141942 | MGC141968 | Nbla04300 | syntaxin binding protein 5 (tomosyn) | Reconstituted Complex Two-hybrid | BioGRID | 12832401 |
| STXBP6 | FLJ39638 | HSPC156 | amisyn | syntaxin binding protein 6 (amisyn) | - | HPRD | 12145319 |
| SYT1 | DKFZp781D2042 | P65 | SVP65 | SYT | synaptotagmin I | Reconstituted Complex | BioGRID | 10397765 |
| SYT4 | HsT1192 | KIAA1342 | synaptotagmin IV | - | HPRD | 10397765 |
| SYT7 | IPCA-7 | MGC150517 | PCANAP7 | SYT-VII | synaptotagmin VII | - | HPRD | 7791877 |
| TXLNA | DKFZp451J0118 | IL14 | MGC118870 | MGC118871 | RP4-622L5.4 | TXLN | taxilin alpha | Reconstituted Complex | BioGRID | 12558796 |
| TXLNB | C6orf198 | DKFZp451A175 | LST001 | MDP77 | dJ522B19.2 | taxilin beta | - | HPRD,BioGRID | 15184072 |
| VAMP1 | DKFZp686H12131 | SYB1 | VAMP-1 | vesicle-associated membrane protein 1 (synaptobrevin 1) | Two-hybrid | BioGRID | 8760387 |
| VAMP2 | FLJ11460 | SYB2 | VAMP-2 | vesicle-associated membrane protein 2 (synaptobrevin 2) | Affinity Capture-Western Two-hybrid | BioGRID | 8760387 |10194441 |10820264 |12517971 |
| VAMP2 | FLJ11460 | SYB2 | VAMP-2 | vesicle-associated membrane protein 2 (synaptobrevin 2) | - | HPRD | 8760387 |12417022 |12517971|8770861 |
| VAMP3 | CEB | vesicle-associated membrane protein 3 (cellubrevin) | - | HPRD,BioGRID | 12130530 |
| VAMP4 | VAMP24 | vesicle-associated membrane protein 4 | Two-hybrid | BioGRID | 16189514 |
| VAMP7 | SYBL1 | TI-VAMP | VAMP-7 | vesicle-associated membrane protein 7 | Affinity Capture-Western | BioGRID | 12853575 |
| VAMP8 | EDB | vesicle-associated membrane protein 8 (endobrevin) | - | HPRD,BioGRID | 10820264 |
| VAPB | ALS8 | VAMP-B | VAMP-C | VAP-B | VAP-C | VAMP (vesicle-associated membrane protein)-associated protein B and C | Reconstituted Complex | BioGRID | 12651853 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SNARE INTERACTIONS IN VESICULAR TRANSPORT | 38 | 25 | All SZGR 2.0 genes in this pathway |
| KEGG VASOPRESSIN REGULATED WATER REABSORPTION | 44 | 30 | All SZGR 2.0 genes in this pathway |
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| PID INSULIN GLUCOSE PATHWAY | 26 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME MEMBRANE TRAFFICKING | 129 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME BOTULINUM NEUROTOXICITY | 19 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | 60 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP | 244 | 165 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS | 47 | 28 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS MODEL UP | 45 | 26 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE VIA P38 COMPLETE | 227 | 151 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-199 | 104 | 110 | m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.