Gene Page: ZNF142
Summary ?
| GeneID | 7701 |
| Symbol | ZNF142 |
| Synonyms | pHZ-49 |
| Description | zinc finger protein 142 |
| Reference | MIM:604083|HGNC:HGNC:12927|Ensembl:ENSG00000115568|HPRD:04967|Vega:OTTHUMG00000154736 |
| Gene type | protein-coding |
| Map location | 2q35 |
| Pascal p-value | 0.661 |
| Sherlock p-value | 0.068 |
| Fetal beta | 0.419 |
| DMG | 1 (# studies) |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01016 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00916 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05372499 | 2 | 219523924 | ZNF142 | 8.17E-9 | -0.009 | 3.87E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
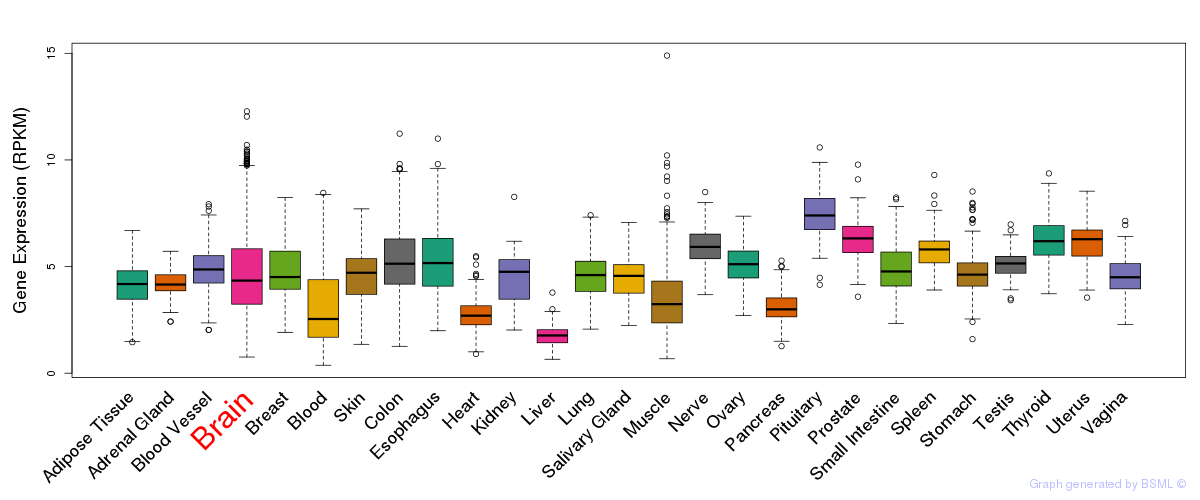
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KDM5A | 0.97 | 0.96 |
| ARID2 | 0.96 | 0.95 |
| BTBD7 | 0.96 | 0.95 |
| PBRM1 | 0.95 | 0.92 |
| CHD9 | 0.95 | 0.95 |
| PAK2 | 0.95 | 0.93 |
| MYST3 | 0.95 | 0.94 |
| PRRC1 | 0.95 | 0.91 |
| MED13 | 0.95 | 0.94 |
| PPP2R3A | 0.95 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.64 | -0.69 |
| AF347015.31 | -0.64 | -0.80 |
| IFI27 | -0.64 | -0.81 |
| HLA-F | -0.63 | -0.67 |
| PTH1R | -0.63 | -0.68 |
| MT-CO2 | -0.63 | -0.80 |
| FXYD1 | -0.63 | -0.79 |
| AF347015.27 | -0.62 | -0.76 |
| AIFM3 | -0.62 | -0.67 |
| TNFSF12 | -0.62 | -0.60 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 7557990 | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0009055 | electron carrier activity | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |