Gene Page: ABHD16A
Summary ?
| GeneID | 7920 |
| Symbol | ABHD16A |
| Synonyms | BAT5|D6S82E|NG26|PP199 |
| Description | abhydrolase domain containing 16A |
| Reference | MIM:142620|HGNC:HGNC:13921|Ensembl:ENSG00000204427|HPRD:15929|Vega:OTTHUMG00000031177 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 1E-12 |
| Sherlock p-value | 0.101 |
| Fetal beta | -0.237 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14986464 | 6 | 31669902 | ABHD16A | 2.6E-4 | -0.005 | 0.172 | DMG:Montano_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2518027 | chr6 | 31436475 | ABHD16A | 7920 | 0.17 | cis |
Section II. Transcriptome annotation
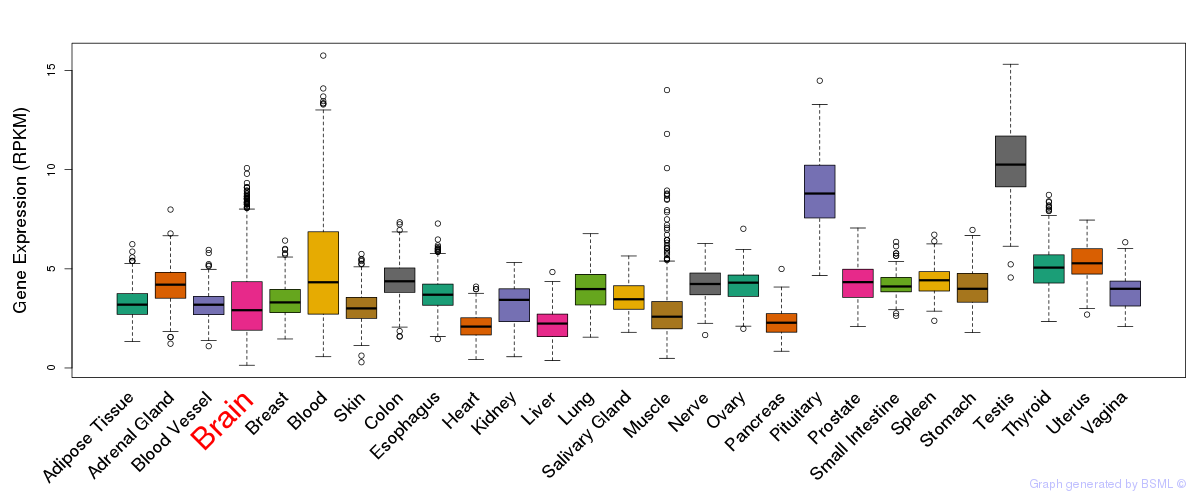
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 14667819 |15231747 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATP5G3 | MGC125738 | P3 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) | - | HPRD,BioGRID | 14667819 |
| DNAJC1 | DNAJL1 | ERdj1 | HTJ1 | MGC131954 | MTJ1 | DnaJ (Hsp40) homolog, subfamily C, member 1 | - | HPRD,BioGRID | 14667819 |
| GPRC5C | MGC131820 | RAIG-3 | RAIG3 | G protein-coupled receptor, family C, group 5, member C | - | HPRD,BioGRID | 14667819 |
| HM13 | H13 | IMP1 | IMPAS | MSTP086 | PSENL3 | PSL3 | SPP | dJ324O17.1 | histocompatibility (minor) 13 | - | HPRD,BioGRID | 14667819 |
| HNRNPA1 | HNRPA1 | MGC102835 | heterogeneous nuclear ribonucleoprotein A1 | - | HPRD | 14667819 |
| IFITM1 | 9-27 | CD225 | IFI17 | LEU13 | interferon induced transmembrane protein 1 (9-27) | - | HPRD,BioGRID | 14667819 |
| PILRB | FDFACT1 | FDFACT2 | paired immunoglobin-like type 2 receptor beta | - | HPRD | 14667819 |
| PTGS2 | COX-2 | COX2 | GRIPGHS | PGG/HS | PGHS-2 | PHS-2 | hCox-2 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) | - | HPRD | 14667819 |
| RNF5 | RING5 | RMA1 | ring finger protein 5 | - | HPRD,BioGRID | 14667819 |
| SAFB | DKFZp779C1727 | HAP | HET | SAFB1 | scaffold attachment factor B | - | HPRD,BioGRID | 14667819 |
| SPAG7 | ACRP | FSA-1 | MGC20134 | sperm associated antigen 7 | - | HPRD,BioGRID | 14667819 |
| TMEM147 | MGC1936 | NIFIE14 | transmembrane protein 147 | - | HPRD,BioGRID | 14667819 |
| TMEM222 | C1orf160 | DKFZp564D0478 | MGC111002 | transmembrane protein 222 | - | HPRD,BioGRID | 14667819 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PETRETTO HEART MASS QTL CIS UP | 28 | 15 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |