Gene Page: GAL3ST4
Summary ?
| GeneID | 79690 |
| Symbol | GAL3ST4 |
| Synonyms | GAL3ST-4 |
| Description | galactose-3-O-sulfotransferase 4 |
| Reference | MIM:608235|HGNC:HGNC:24145|Ensembl:ENSG00000197093|HPRD:12195|Vega:OTTHUMG00000154885 |
| Gene type | protein-coding |
| Map location | 7q22.1 |
| Pascal p-value | 0.135 |
| Fetal beta | 2.483 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Expression | Meta-analysis of gene expression | P value: 1.58 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08398147 | 7 | 99766376 | GAL3ST4 | 3.216E-4 | -0.248 | 0.04 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12785310 | chr11 | 122601618 | GAL3ST4 | 79690 | 0.18 | trans |
Section II. Transcriptome annotation
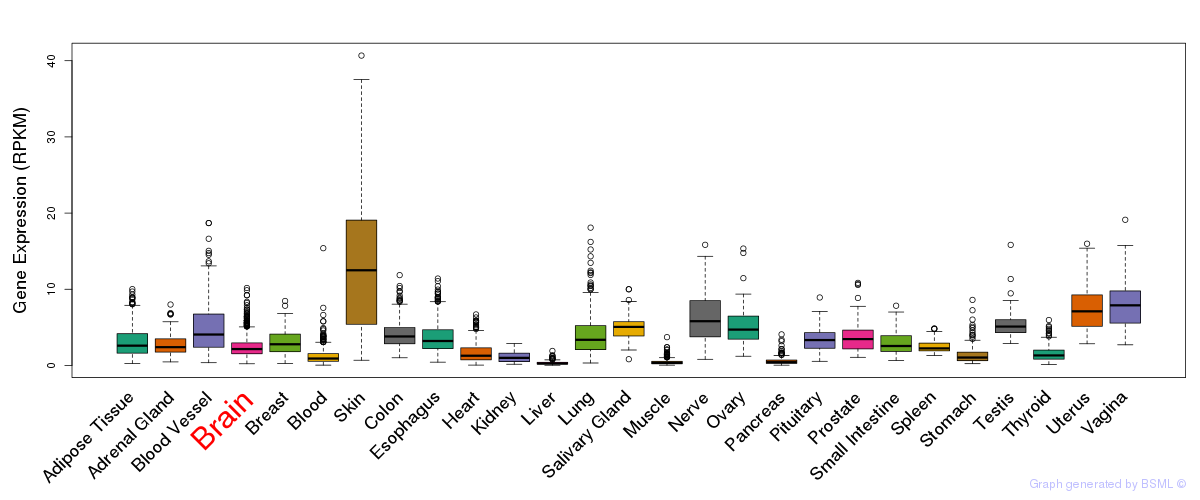
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001733 | galactosylceramide sulfotransferase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding | NAS | 11333265 | |
| GO:0050698 | proteoglycan sulfotransferase activity | NAS | 11333265 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0009100 | glycoprotein metabolic process | NAS | 11333265 | |
| GO:0007267 | cell-cell signaling | NAS | 11333265 | |
| GO:0030166 | proteoglycan biosynthetic process | NAS | 11333265 | |
| GO:0009311 | oligosaccharide metabolic process | NAS | 11333265 | |
| GO:0009058 | biosynthetic process | IEA | - | |
| GO:0006790 | sulfur metabolic process | NAS | 11333265 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005794 | Golgi apparatus | IEA | - | |
| GO:0005624 | membrane fraction | IDA | 11333265 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | NAS | 11333265 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP | 544 | 308 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS E UP | 97 | 60 | All SZGR 2.0 genes in this pathway |
| WHITEHURST PACLITAXEL SENSITIVITY | 39 | 19 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K4ME3 | 128 | 68 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K27ME3 | 10 | 7 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC LCP WITH H3K4ME3 | 58 | 34 | All SZGR 2.0 genes in this pathway |