Gene Page: THAP7
Summary ?
| GeneID | 80764 |
| Symbol | THAP7 |
| Synonyms | - |
| Description | THAP domain containing 7 |
| Reference | MIM:609518|HGNC:HGNC:23190|Ensembl:ENSG00000184436|HPRD:15500|Vega:OTTHUMG00000150879 |
| Gene type | protein-coding |
| Map location | 22q11.2 |
| Pascal p-value | 0.338 |
| Sherlock p-value | 0.457 |
| Fetal beta | -0.032 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22031629 | 22 | 21355869 | THAP7 | 8.03E-8 | -0.012 | 1.88E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1405218 | chr2 | 150581361 | THAP7 | 80764 | 0.16 | trans | ||
| rs11966012 | chr6 | 147914230 | THAP7 | 80764 | 0.15 | trans | ||
| rs17235614 | chr13 | 97953192 | THAP7 | 80764 | 0.01 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
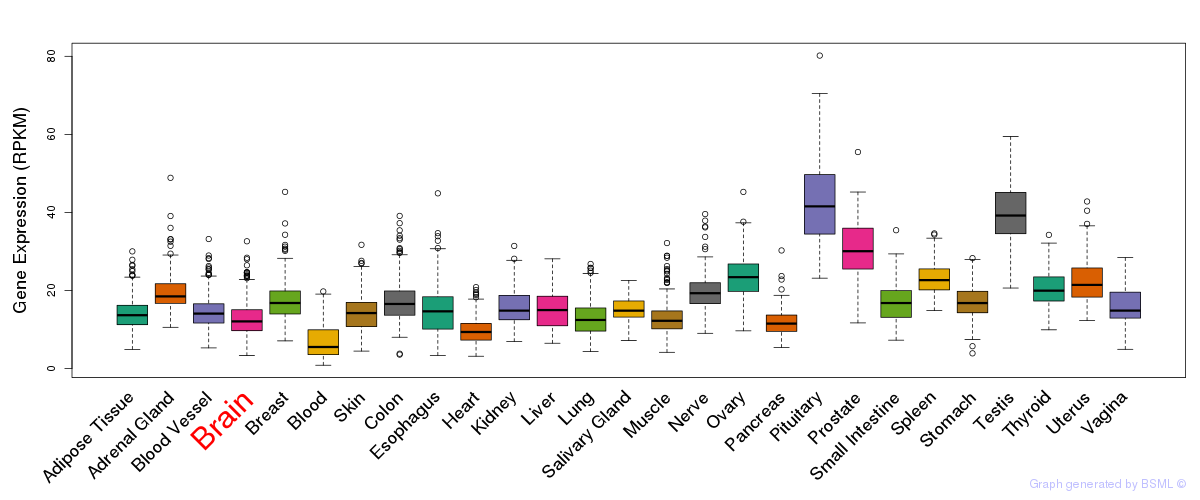
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AIM1L | 0.66 | 0.51 |
| MUC3A | 0.66 | 0.52 |
| C19orf15 | 0.66 | 0.57 |
| POMT1 | 0.65 | 0.59 |
| HOOK2 | 0.64 | 0.58 |
| KIAA1683 | 0.64 | 0.54 |
| FAM113A | 0.64 | 0.61 |
| WDR85 | 0.63 | 0.62 |
| NAGK | 0.63 | 0.61 |
| QTRT1 | 0.63 | 0.64 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.46 | -0.52 |
| MT-CO2 | -0.45 | -0.50 |
| MT-CYB | -0.45 | -0.51 |
| AF347015.27 | -0.44 | -0.51 |
| AF347015.8 | -0.43 | -0.50 |
| AF347015.33 | -0.43 | -0.48 |
| AF347015.15 | -0.43 | -0.51 |
| ABCG2 | -0.42 | -0.45 |
| AF347015.2 | -0.42 | -0.49 |
| AF347015.9 | -0.42 | -0.50 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003677 | DNA binding | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005694 | chromosome | IEA | - | |
| GO:0031965 | nuclear membrane | IDA | 18029348 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |