Gene Page: RSPH6A
Summary ?
| GeneID | 81492 |
| Symbol | RSPH6A |
| Synonyms | RSHL1|RSP4|RSP6|RSPH4B |
| Description | radial spoke head 6 homolog A (Chlamydomonas) |
| Reference | MIM:607548|HGNC:HGNC:14241|Ensembl:ENSG00000104941|HPRD:09611|Vega:OTTHUMG00000182482 |
| Gene type | protein-coding |
| Map location | 19q13.3 |
| Pascal p-value | 0.217 |
| Fetal beta | -0.128 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17929770 | 19 | 46318514 | RSPH6A | 5.73E-5 | 0.381 | 0.022 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
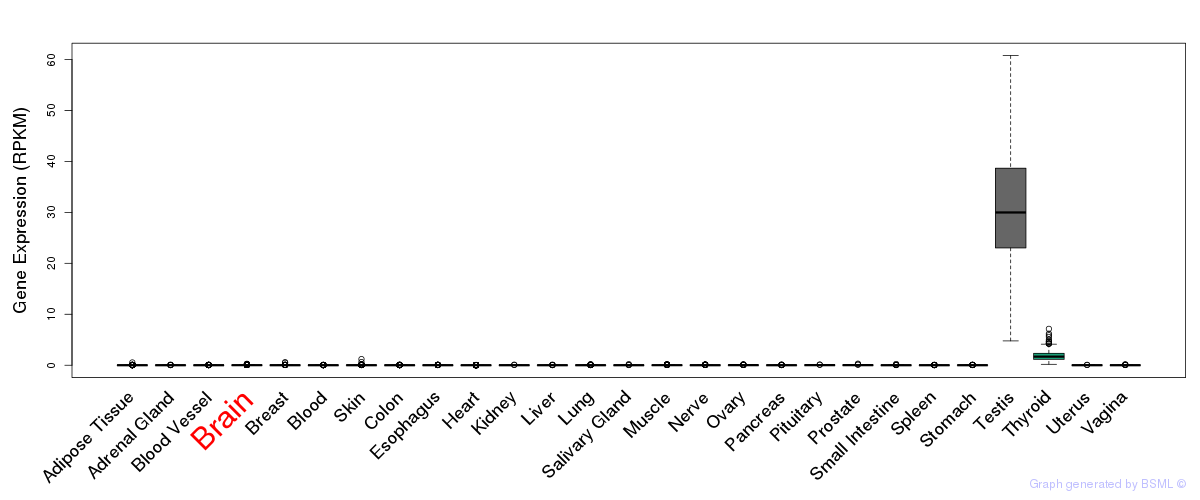
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CCDC84 | 0.56 | 0.46 |
| NPEPL1 | 0.55 | 0.47 |
| ZNF692 | 0.55 | 0.46 |
| LENG8 | 0.55 | 0.46 |
| GDPD3 | 0.54 | 0.44 |
| TSSK3 | 0.54 | 0.44 |
| AGER | 0.54 | 0.45 |
| TNFRSF25 | 0.53 | 0.45 |
| PILRB | 0.53 | 0.46 |
| NEIL1 | 0.53 | 0.45 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CYCS | -0.37 | -0.40 |
| ARL1 | -0.37 | -0.41 |
| PIGK | -0.35 | -0.39 |
| DNAJB9 | -0.35 | -0.42 |
| TPMT | -0.35 | -0.39 |
| DNAJB14 | -0.34 | -0.37 |
| IDH3A | -0.34 | -0.37 |
| GHITM | -0.34 | -0.38 |
| CCDC25 | -0.34 | -0.33 |
| OSGIN2 | -0.33 | -0.38 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008199 | ferric iron binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006826 | iron ion transport | IEA | - | |
| GO:0006879 | cellular iron ion homeostasis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005622 | intracellular | IDA | 11256614 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |