Gene Page: CAPNS1
Summary ?
| GeneID | 826 |
| Symbol | CAPNS1 |
| Synonyms | CALPAIN4|CANP|CANPS|CAPN4|CDPS|CSS1 |
| Description | calpain small subunit 1 |
| Reference | MIM:114170|HGNC:HGNC:1481|Ensembl:ENSG00000126247|HPRD:00240|Vega:OTTHUMG00000160811 |
| Gene type | protein-coding |
| Map location | 19q13.12 |
| Pascal p-value | 8.764E-4 |
| Sherlock p-value | 0.969 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=-1.09:CC_BA10_disease_P=0.0323:HBB_BA9_fold_change=-1.19:HBB_BA9_disease_P=0.0420 |
| Fetal beta | -1.19 |
| DMG | 1 (# studies) |
| eGene | Putamen basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20717845 | 19 | 36630602 | CAPNS1 | 1.48E-8 | -0.008 | 5.66E-6 | DMG:Jaffe_2016 |
| cg19372504 | 19 | 36633213 | CAPNS1 | 5.98E-8 | 0.018 | 1.51E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs9966136 | chr18 | 74881659 | CAPNS1 | 826 | 0.12 | trans | ||
| rs139322373 | 19 | 36823631 | CAPNS1 | ENSG00000126247.6 | 2.94415E-6 | 0.05 | 193154 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
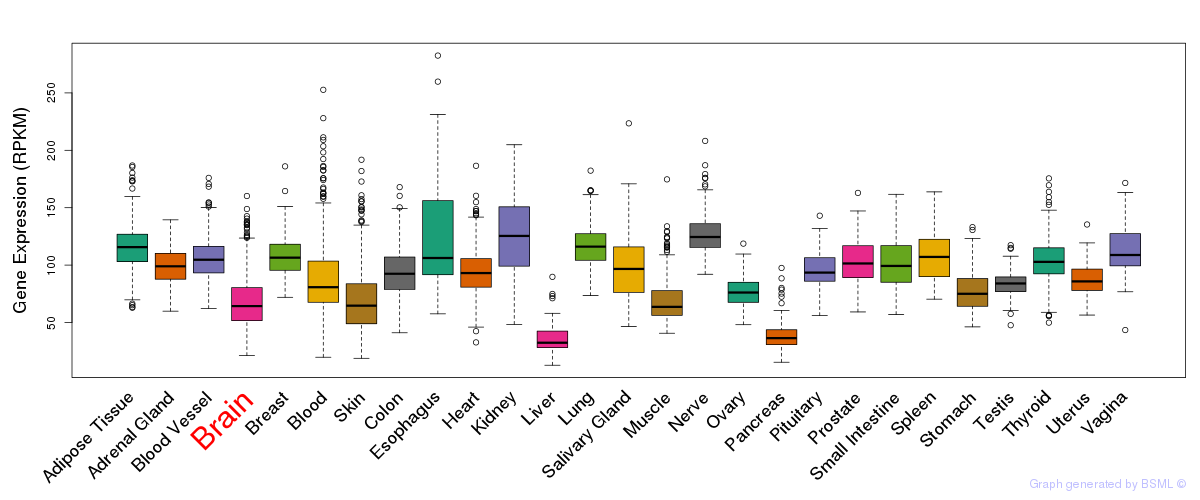
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CHMP5 | 0.87 | 0.86 |
| TXNDC9 | 0.87 | 0.85 |
| TMEM49 | 0.86 | 0.88 |
| COPS4 | 0.86 | 0.87 |
| CCNC | 0.85 | 0.84 |
| OLA1 | 0.84 | 0.87 |
| ACTR6 | 0.84 | 0.84 |
| COMMD10 | 0.83 | 0.82 |
| MRPS35 | 0.83 | 0.84 |
| COPS2 | 0.82 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GPT | -0.56 | -0.63 |
| MICALL2 | -0.55 | -0.64 |
| FAM38A | -0.54 | -0.57 |
| TENC1 | -0.53 | -0.55 |
| AF347015.26 | -0.52 | -0.53 |
| C1orf175 | -0.52 | -0.60 |
| SLC27A1 | -0.52 | -0.57 |
| AC010618.2 | -0.52 | -0.58 |
| DOCK6 | -0.50 | -0.52 |
| PPP1R13L | -0.50 | -0.50 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARR3 | ARRX | arrestin 3, retinal (X-arrestin) | Two-hybrid | BioGRID | 16169070 |
| ATP5J2 | ATP5JL | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 | Two-hybrid | BioGRID | 16169070 |
| C7orf20 | CEE | CGI-20 | chromosome 7 open reading frame 20 | Two-hybrid | BioGRID | 16169070 |
| CAPN1 | CANP | CANP1 | CANPL1 | muCANP | muCL | calpain 1, (mu/I) large subunit | - | HPRD | 12591934 |
| CAPN1 | CANP | CANP1 | CANPL1 | muCANP | muCL | calpain 1, (mu/I) large subunit | CAPN1 interacts with CAPNS1. | BIND | 12358155 |
| CAPN2 | CANP2 | CANPL2 | CANPml | FLJ39928 | mCANP | calpain 2, (m/II) large subunit | - | HPRD,BioGRID | 10639123 |10666632 |11517928 |11673052 |
| CAST | BS-17 | MGC9402 | calpastatin | - | HPRD | 12684003 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | Two-hybrid | BioGRID | 16169070 |
| DRG1 | DKFZp434N1827 | NEDD3 | developmentally regulated GTP binding protein 1 | Two-hybrid | BioGRID | 16169070 |
| GNB2 | - | guanine nucleotide binding protein (G protein), beta polypeptide 2 | Two-hybrid | BioGRID | 16169070 |
| IL2RG | CD132 | IMD4 | SCIDX | SCIDX1 | interleukin 2 receptor, gamma (severe combined immunodeficiency) | - | HPRD,BioGRID | 9326644 |
| RAB1A | DKFZp564B163 | RAB1 | YPT1 | RAB1A, member RAS oncogene family | Two-hybrid | BioGRID | 16169070 |
| TERF1 | FLJ41416 | PIN2 | TRBF1 | TRF | TRF1 | hTRF1-AS | t-TRF1 | telomeric repeat binding factor (NIMA-interacting) 1 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA P35ALZHEIMERS PATHWAY | 11 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MCALPAIN PATHWAY | 25 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MEF2D PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA UCALPAIN PATHWAY | 18 | 11 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL DN | 101 | 66 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING DN | 90 | 57 | All SZGR 2.0 genes in this pathway |
| PARK HSC VS MULTIPOTENT PROGENITORS UP | 19 | 14 | All SZGR 2.0 genes in this pathway |
| ZHAN V2 LATE DIFFERENTIATION GENES | 45 | 34 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS RESPONSE | 35 | 28 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G4 | 21 | 15 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL DN | 102 | 65 | All SZGR 2.0 genes in this pathway |
| NUTT GBM VS AO GLIOMA UP | 46 | 34 | All SZGR 2.0 genes in this pathway |
| HOFMANN MYELODYSPLASTIC SYNDROM LOW RISK DN | 30 | 20 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 12 | 79 | 54 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| KOHOUTEK CCNT1 TARGETS | 50 | 26 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |