Gene Page: DOC2A
Summary ?
| GeneID | 8448 |
| Symbol | DOC2A |
| Synonyms | Doc2 |
| Description | double C2-like domains, alpha |
| Reference | MIM:604567|HGNC:HGNC:2985|Ensembl:ENSG00000149927|HPRD:05195|Vega:OTTHUMG00000132109 |
| Gene type | protein-coding |
| Map location | 16p11.2 |
| Pascal p-value | 7.684E-9 |
| Sherlock p-value | 0.953 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=-1.12:CC_BA10_disease_P=0.0497:HBB_BA9_fold_change=-1.30:HBB_BA9_disease_P=0.0225 |
| Fetal beta | -0.765 |
| Support | EXOCYTOSIS G2Cdb.humanPSP Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 4 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
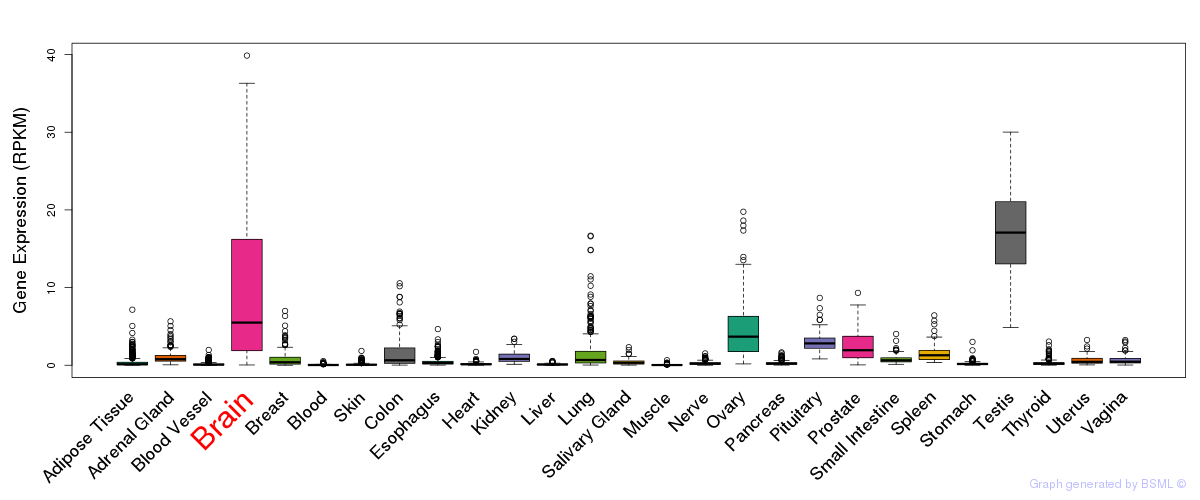
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SYT12 | 0.94 | 0.88 |
| ZFYVE28 | 0.92 | 0.91 |
| LYNX1 | 0.92 | 0.87 |
| CAMKK1 | 0.92 | 0.88 |
| CNTNAP1 | 0.91 | 0.91 |
| IQSEC3 | 0.91 | 0.94 |
| CAMK2A | 0.91 | 0.90 |
| EXTL1 | 0.91 | 0.88 |
| LGI3 | 0.91 | 0.81 |
| ZBTB7A | 0.91 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.54 | -0.61 |
| C9orf46 | -0.51 | -0.58 |
| TUBB2B | -0.51 | -0.52 |
| DYNLT1 | -0.50 | -0.61 |
| KIAA1949 | -0.50 | -0.39 |
| GTF3C6 | -0.49 | -0.50 |
| TRAF4 | -0.49 | -0.51 |
| YBX1 | -0.49 | -0.44 |
| PDE9A | -0.49 | -0.48 |
| SH3BP2 | -0.49 | -0.50 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005544 | calcium-dependent phospholipid binding | IEA | - | |
| GO:0005488 | binding | TAS | 7826360 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005215 | transporter activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 7826360 |
| GO:0007399 | nervous system development | TAS | neurite (GO term level: 5) | 7826360 |
| GO:0006810 | transport | IEA | - | |
| GO:0006887 | exocytosis | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008021 | synaptic vesicle | IEA | Synap, Neurotransmitter (GO term level: 12) | - |
| GO:0019717 | synaptosome | IEA | Synap, Brain (GO term level: 7) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0016020 | membrane | IEA | - | |
| GO:0030054 | cell junction | IEA | - | |
| GO:0031410 | cytoplasmic vesicle | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| MUELLER COMMON TARGETS OF AML FUSIONS DN | 31 | 21 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-153 | 216 | 222 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| hsa-miR-153 | UUGCAUAGUCACAAAAGUGA | ||||
| miR-196 | 105 | 112 | 1A,m8 | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-30-5p | 74 | 80 | 1A | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG | ||||
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-448 | 177 | 184 | 1A,m8 | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU | ||||
| miR-452 | 180 | 186 | m8 | hsa-miR-452 | UGUUUGCAGAGGAAACUGAGAC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.