Gene Page: CASK
Summary ?
| GeneID | 8573 |
| Symbol | CASK |
| Synonyms | CAGH39|CAMGUK|CMG|FGS4|LIN2|MICPCH|MRXSNA|TNRC8 |
| Description | calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| Reference | MIM:300172|HGNC:HGNC:1497|Ensembl:ENSG00000147044|HPRD:02164|Vega:OTTHUMG00000021378 |
| Gene type | protein-coding |
| Map location | Xp11.4 |
| Fetal beta | 1.432 |
| Support | PROTEIN CLUSTERING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS Ascano FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 1.461 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1861 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
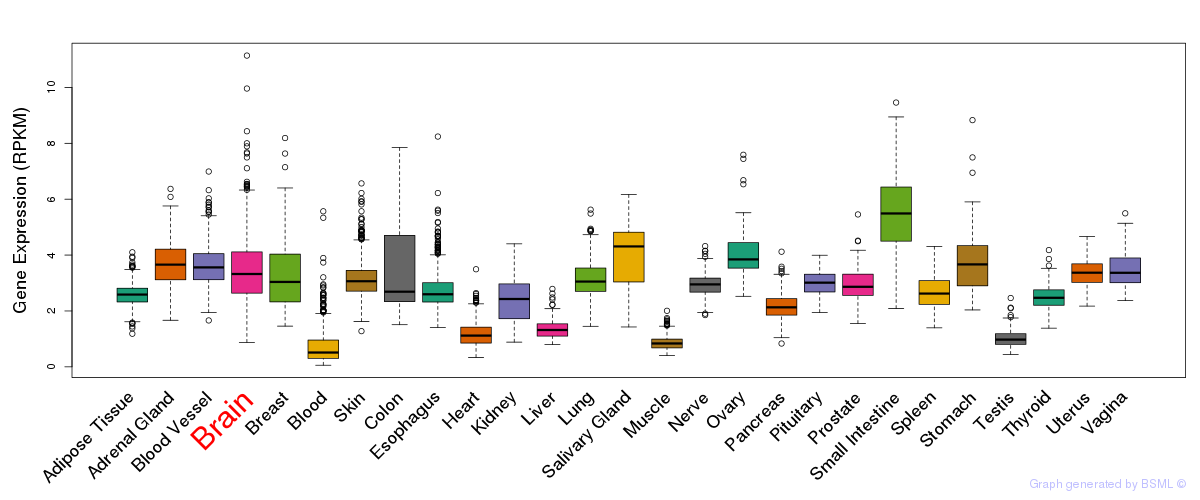
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1305 | 0.97 | 0.95 |
| PLXNB2 | 0.97 | 0.97 |
| PLEKHG2 | 0.97 | 0.92 |
| PLXNA3 | 0.97 | 0.93 |
| KDM6B | 0.95 | 0.93 |
| GTF2IRD1 | 0.95 | 0.89 |
| CELSR3 | 0.94 | 0.84 |
| AD000671.1 | 0.94 | 0.89 |
| CASP2 | 0.94 | 0.85 |
| ZNF646 | 0.94 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.69 | -0.81 |
| FBXO2 | -0.67 | -0.65 |
| HLA-F | -0.67 | -0.69 |
| ACOT13 | -0.66 | -0.77 |
| ALDOC | -0.66 | -0.64 |
| ACYP2 | -0.65 | -0.73 |
| S100B | -0.65 | -0.77 |
| AIFM3 | -0.65 | -0.63 |
| CA4 | -0.65 | -0.76 |
| LDHD | -0.65 | -0.61 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005516 | calmodulin binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004385 | guanylate kinase activity | TAS | 9660868 | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | TAS | 9660868 | |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | TAS | 9660868 | |
| GO:0015629 | actin cytoskeleton | TAS | 9660868 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APBA1 | D9S411E | MINT1 | X11 | X11A | X11ALPHA | amyloid beta (A4) precursor protein-binding, family A, member 1 | X11-ALPHA interacts with Lin-2. | BIND | 9822620 |
| APBA1 | D9S411E | MINT1 | X11 | X11A | X11ALPHA | amyloid beta (A4) precursor protein-binding, family A, member 1 | - | HPRD,BioGRID | 9753324 |9822620 |9952408|9822620 |9952408 |
| ATP2B4 | ATP2B2 | DKFZp686G08106 | DKFZp686M088 | MXRA1 | PMCA4 | PMCA4b | PMCA4x | ATPase, Ca++ transporting, plasma membrane 4 | - | HPRD,BioGRID | 12511555 |
| C16orf70 | C16orf6 | FLJ12076 | LIN10 | lin-10 | chromosome 16 open reading frame 70 | Reconstituted Complex | BioGRID | 9822620 |
| CACNA1B | BIII | CACNL1A5 | CACNN | Cav2.2 | calcium channel, voltage-dependent, N type, alpha 1B subunit | The SH3 domain of CASK interacts with the carboxy-terminal proline-rich region (PRR) of Ca-alpha-1B. | BIND | 10455105 |
| CADM1 | BL2 | DKFZp686F1789 | IGSF4 | IGSF4A | MGC149785 | MGC51880 | NECL2 | Necl-2 | RA175 | ST17 | SYNCAM | TSLC1 | sTSLC-1 | sgIGSF | synCAM1 | cell adhesion molecule 1 | - | HPRD,BioGRID | 12202822 |
| CASKIN1 | ANKS5A | CASK interacting protein 1 | - | HPRD | 12040031 |
| CD2AP | CMS | DKFZp586H0519 | CD2-associated protein | Reconstituted Complex | BioGRID | 15331416 |
| CNTNAP2 | AUTS15 | CASPR2 | CDFE | DKFZp781D1846 | NRXN4 | contactin associated protein-like 2 | - | HPRD,BioGRID | 12093160 |
| CNTNAP4 | CASPR4 | KIAA1763 | contactin associated protein-like 4 | - | HPRD,BioGRID | 12093160 |
| DFNB31 | CIP98 | DKFZp434N014 | KIAA1526 | RP11-9M16.1 | USH2D | WHRN | WI | deafness, autosomal recessive 31 | - | HPRD,BioGRID | 12641734 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | Affinity Capture-Western in vitro in vivo Reconstituted Complex Two-hybrid | BioGRID | 10993877 |12151521 |12351654 |14960569 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | CASK interacts with DLG1 (SAP97). | BIND | 15729360 |
| DLG1 | DKFZp761P0818 | DKFZp781B0426 | DLGH1 | SAP97 | dJ1061C18.1.1 | hdlg | discs, large homolog 1 (Drosophila) | - | HPRD | 10993877 |12351654 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD,BioGRID | 12151521 |
| EPB41 | 4.1R | EL1 | HE | erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | - | HPRD,BioGRID | 9660868 |
| F11R | CD321 | JAM | JAM-1 | JAM-A | JAM1 | JAMA | JCAM | KAT | PAM-1 | F11 receptor | - | HPRD,BioGRID | 11120739 |
| FCHSD2 | KIAA0769 | NWK | SH3MD3 | FCH and double SH3 domains 2 | - | HPRD,BioGRID | 14627983 |
| GRIK2 | EAA4 | GLR6 | GLUK6 | GLUR6 | MGC74427 | MRT6 | glutamate receptor, ionotropic, kainate 2 | - | HPRD | 12151522 |
| ID1 | ID | bHLHb24 | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | CASK interacts with Id1. | BIND | 15694377 |
| ID1 | ID | bHLHb24 | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | CASK interacts with Id1'. | BIND | 15694377 |
| ID1 | ID | bHLHb24 | inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | Reconstituted Complex Two-hybrid | BioGRID | 15694377 |
| KCNJ12 | FLJ14167 | IRK2 | KCNJN1 | Kir2.2 | Kir2.2v | hIRK | hIRK1 | hkir2.2x | kcnj12x | potassium inwardly-rectifying channel, subfamily J, member 12 | - | HPRD,BioGRID | 14960569 |15024025 |
| KCNJ4 | HIR | HIRK2 | HRK1 | IRK3 | Kir2.3 | MGC142066 | MGC142068 | potassium inwardly-rectifying channel, subfamily J, member 4 | - | HPRD | 11742811 |
| KCNJ4 | HIR | HIRK2 | HRK1 | IRK3 | Kir2.3 | MGC142066 | MGC142068 | potassium inwardly-rectifying channel, subfamily J, member 4 | Affinity Capture-Western | BioGRID | 14960569 |15024025 |
| LIN7A | LIN-7A | LIN7 | MALS-1 | MGC148143 | TIP-33 | VELI1 | lin-7 homolog A (C. elegans) | Affinity Capture-Western in vitro in vivo Reconstituted Complex | BioGRID | 9753324 |9822620 |14960569 |
| LIN7A | LIN-7A | LIN7 | MALS-1 | MGC148143 | TIP-33 | VELI1 | lin-7 homolog A (C. elegans) | - | HPRD | 9753324|9822620 |9952408 |
| LIN7B | LIN-7B | MALS-2 | MALS2 | VELI2 | lin-7 homolog B (C. elegans) | - | HPRD | 11742811 |
| LIN7B | LIN-7B | MALS-2 | MALS2 | VELI2 | lin-7 homolog B (C. elegans) | Affinity Capture-Western | BioGRID | 14960569 |
| LIN7C | FLJ11215 | LIN-7-C | LIN-7C | MALS-3 | MALS3 | VELI3 | lin-7 homolog C (C. elegans) | Affinity Capture-Western | BioGRID | 14960569 |
| NPHS1 | CNF | NPHN | nephrosis 1, congenital, Finnish type (nephrin) | Affinity Capture-MS Reconstituted Complex | BioGRID | 15331416 |
| NRXN1 | DKFZp313P2036 | FLJ35941 | Hs.22998 | KIAA0578 | neurexin 1 | - | HPRD,BioGRID | 11036064 |
| NRXN1 | DKFZp313P2036 | FLJ35941 | Hs.22998 | KIAA0578 | neurexin 1 | - | HPRD | 8786425 |
| NRXN2 | FLJ40892 | KIAA0921 | neurexin 2 | - | HPRD | 8786425 |
| NRXN3 | KIAA0743 | MGC176711 | neurexin 3 | - | HPRD,BioGRID | 8786425 |
| PARK2 | AR-JP | LPRS2 | PDJ | PRKN | Parkinson disease (autosomal recessive, juvenile) 2, parkin | - | HPRD,BioGRID | 11679592 |
| RAB3A | - | RAB3A, member RAS oncogene family | - | HPRD | 11377421 |
| RPH3A | KIAA0985 | rabphilin 3A homolog (mouse) | - | HPRD,BioGRID | 11377421 |
| SDC1 | CD138 | SDC | SYND1 | syndecan | syndecan 1 | - | HPRD,BioGRID | 9660868 |
| SDC2 | HSPG | HSPG1 | SYND2 | syndecan 2 | - | HPRD,BioGRID | 9660868 |
| SDC3 | N-syndecan | SDCN | SYND3 | syndecan 3 | - | HPRD | 10460248 |
| TANC1 | KIAA1728 | ROLSB | TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | - | HPRD | 15673434 |
| TBR1 | MGC141978 | TES-56 | T-box, brain, 1 | - | HPRD,BioGRID | 10749215 |
| TSPYL2 | CDA1 | CINAP | CTCL | DENTT | HRIHFB2216 | SE20-4 | TSPY-like 2 | Affinity Capture-Western Two-hybrid | BioGRID | 15066269 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 1 PATHWAY | 46 | 29 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 2 PATHWAY | 33 | 27 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 3 PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS UP | 501 | 327 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR UP | 162 | 116 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 UP | 137 | 84 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION KRAS UP | 126 | 72 | All SZGR 2.0 genes in this pathway |
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 UP | 120 | 73 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA UP | 207 | 145 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 AND CD2 UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| WEIGEL OXIDATIVE STRESS RESPONSE | 35 | 28 | All SZGR 2.0 genes in this pathway |
| ULE SPLICING VIA NOVA2 | 43 | 38 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 10HR UP | 101 | 69 | All SZGR 2.0 genes in this pathway |
| WANG SMARCE1 TARGETS UP | 280 | 183 | All SZGR 2.0 genes in this pathway |
| RIGGINS TAMOXIFEN RESISTANCE DN | 220 | 147 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D DN | 78 | 34 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| SYED ESTRADIOL RESPONSE | 19 | 15 | All SZGR 2.0 genes in this pathway |
| PYEON CANCER HEAD AND NECK VS CERVICAL UP | 193 | 95 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN UP | 56 | 38 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR UP | 55 | 41 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-146 | 207 | 213 | m8 | hsa-miR-146a | UGAGAACUGAAUUCCAUGGGUU |
| hsa-miR-146bbrain | UGAGAACUGAAUUCCAUAGGCU | ||||
| miR-196 | 98 | 105 | 1A,m8 | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-369-3p | 247 | 253 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-370 | 76 | 82 | m8 | hsa-miR-370brain | GCCUGCUGGGGUGGAACCUGG |
| miR-410 | 269 | 275 | m8 | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-495 | 57 | 63 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-543 | 265 | 272 | 1A,m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.