Gene Page: NOP14
Summary ?
| GeneID | 8602 |
| Symbol | NOP14 |
| Synonyms | C4orf9|NOL14|RES4-25|RES425|UTP2 |
| Description | NOP14 nucleolar protein |
| Reference | MIM:611526|HGNC:HGNC:16821|Ensembl:ENSG00000087269|HPRD:12827|Vega:OTTHUMG00000159911 |
| Gene type | protein-coding |
| Map location | 4p16.3 |
| Pascal p-value | 0.046 |
| Sherlock p-value | 0.444 |
| Fetal beta | -0.155 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| Expression | Meta-analysis of gene expression | P value: 1.402 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17695145 | 4 | 2940996 | C4orf10;NOP14 | 4.608E-4 | 0.377 | 0.046 | DMG:Wockner_2014 |
| cg07945480 | 4 | 2964972 | NOP14 | 1.77E-10 | -0.012 | 5.27E-7 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | NOP14 | 8602 | 3.121E-4 | trans | ||
| rs3845734 | chr2 | 171125572 | NOP14 | 8602 | 0.03 | trans | ||
| rs7584986 | chr2 | 184111432 | NOP14 | 8602 | 0.02 | trans | ||
| rs7787830 | chr7 | 98797019 | NOP14 | 8602 | 0.16 | trans | ||
| rs3118341 | chr9 | 25185518 | NOP14 | 8602 | 0.19 | trans | ||
| rs16955618 | chr15 | 29937543 | NOP14 | 8602 | 3.664E-7 | trans | ||
| rs1041786 | chr21 | 22617710 | NOP14 | 8602 | 0.07 | trans |
Section II. Transcriptome annotation
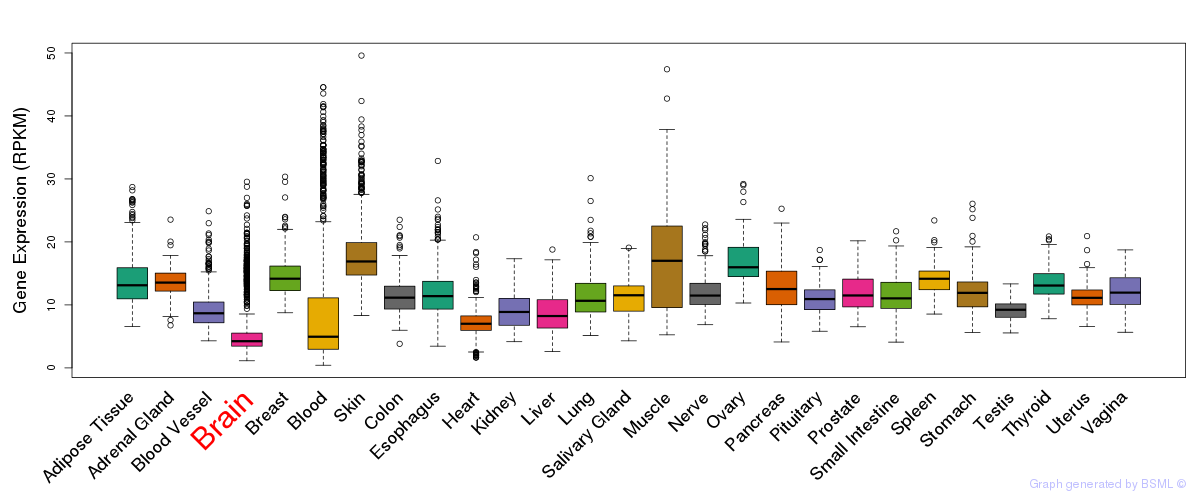
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC1A3 | 0.92 | 0.88 |
| GPC5 | 0.90 | 0.87 |
| ATP13A4 | 0.90 | 0.87 |
| ACSS3 | 0.89 | 0.85 |
| ATP1A2 | 0.89 | 0.93 |
| LRP4 | 0.88 | 0.84 |
| SLC1A2 | 0.88 | 0.80 |
| ADD3 | 0.87 | 0.89 |
| TLR4 | 0.86 | 0.79 |
| KAT2B | 0.86 | 0.91 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MED19 | -0.45 | -0.55 |
| DCTPP1 | -0.42 | -0.52 |
| DUSP12 | -0.42 | -0.50 |
| POLB | -0.42 | -0.57 |
| RAB33A | -0.41 | -0.54 |
| NR2C2AP | -0.41 | -0.56 |
| STMN2 | -0.41 | -0.46 |
| MPP3 | -0.41 | -0.47 |
| MLLT11 | -0.41 | -0.43 |
| BZW2 | -0.41 | -0.52 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | ISS | - | |
| GO:0030515 | snoRNA binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042274 | ribosomal small subunit biogenesis | ISS | - | |
| GO:0030490 | maturation of SSU-rRNA | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005730 | nucleolus | ISS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| AUNG GASTRIC CANCER | 54 | 20 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER MYC TARGETS AND SERUM RESPONSE DN | 47 | 34 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| KANNAN TP53 TARGETS DN | 21 | 15 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC DN | 228 | 146 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION DN | 98 | 67 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS DN | 292 | 189 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| MUELLER COMMON TARGETS OF AML FUSIONS DN | 31 | 21 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |