Gene Page: HTR3B
Summary ?
| GeneID | 9177 |
| Symbol | HTR3B |
| Synonyms | 5-HT3B |
| Description | 5-hydroxytryptamine receptor 3B |
| Reference | MIM:604654|HGNC:HGNC:5298|Ensembl:ENSG00000149305|HPRD:06875|Vega:OTTHUMG00000168210 |
| Gene type | protein-coding |
| Map location | 11q23.1 |
| Pascal p-value | 0.111 |
| Fetal beta | -0.932 |
| Support | LIGAND GATED ION SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Expression | Meta-analysis of gene expression | P value: 1.521 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 6 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
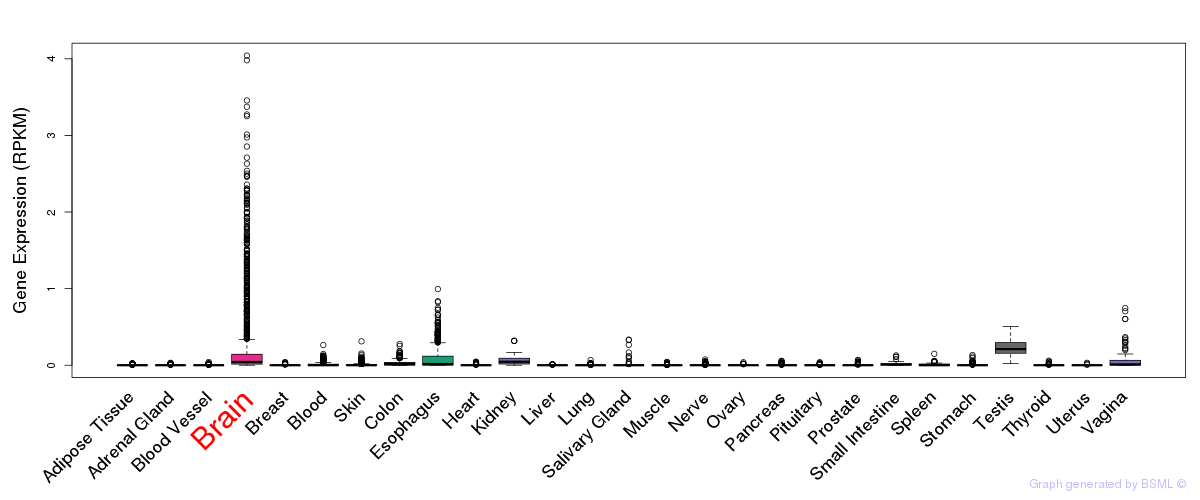
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SCNN1D | 0.82 | 0.84 |
| C16orf79 | 0.76 | 0.78 |
| CPT1B | 0.76 | 0.79 |
| DOM3Z | 0.75 | 0.77 |
| AP1G2 | 0.75 | 0.83 |
| PILRB | 0.75 | 0.79 |
| LIME1 | 0.75 | 0.76 |
| HSF4 | 0.75 | 0.79 |
| NOXA1 | 0.75 | 0.78 |
| MAN2C1 | 0.74 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ABCD2 | -0.37 | -0.33 |
| GMFB | -0.35 | -0.31 |
| PNPLA8 | -0.35 | -0.34 |
| LMBRD2 | -0.35 | -0.29 |
| TTC33 | -0.33 | -0.29 |
| HSPA13 | -0.33 | -0.25 |
| RAB30 | -0.33 | -0.25 |
| MOBKL3 | -0.32 | -0.28 |
| RLIM | -0.32 | -0.25 |
| AGPS | -0.32 | -0.27 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0030594 | neurotransmitter receptor activity | IEA | Neurotransmitter (GO term level: 5) | - |
| GO:0005232 | serotonin-activated cation-selective channel activity | TAS | serotonin (GO term level: 11) | 9950429 |
| GO:0004993 | serotonin receptor activity | TAS | serotonin, Neurotransmitter (GO term level: 8) | 9950429 |
| GO:0005216 | ion channel activity | TAS | 9950429 | |
| GO:0005230 | extracellular ligand-gated ion channel activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 9950429 |
| GO:0007210 | serotonin receptor signaling pathway | IEA | serotonin (GO term level: 8) | - |
| GO:0006811 | ion transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9950429 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| REACTOME ION CHANNEL TRANSPORT | 55 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME LIGAND GATED ION CHANNEL TRANSPORT | 21 | 21 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR DN | 57 | 45 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K27ME3 | 70 | 35 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K27ME3 | 14 | 8 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |