Gene Page: NRXN3
Summary ?
| GeneID | 9369 |
| Symbol | NRXN3 |
| Synonyms | C14orf60 |
| Description | neurexin 3 |
| Reference | MIM:600567|HGNC:HGNC:8010|Ensembl:ENSG00000021645|HPRD:08989|Vega:OTTHUMG00000171502 |
| Gene type | protein-coding |
| Map location | 14q31 |
| Sherlock p-value | 0.224 |
| Fetal beta | -2.391 |
| eGene | Myers' cis & trans |
| Support | CELL ADHESION AND TRANSSYNAPTIC SIGNALING G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | NRXN3 | 9369 | 7.424E-7 | trans | ||
| rs7584986 | chr2 | 184111432 | NRXN3 | 9369 | 0.13 | trans | ||
| rs2393316 | chr10 | 59333070 | NRXN3 | 9369 | 0.19 | trans | ||
| rs16955618 | chr15 | 29937543 | NRXN3 | 9369 | 6.938E-8 | trans |
Section II. Transcriptome annotation
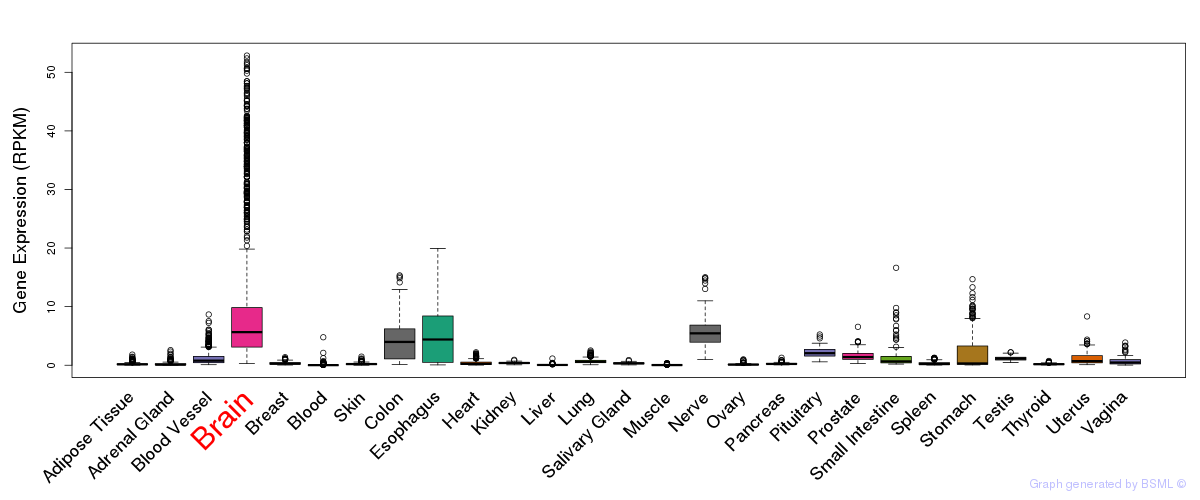
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PADI2 | 0.78 | 0.80 |
| ICAM1 | 0.77 | 0.71 |
| KCNJ10 | 0.76 | 0.76 |
| FOXF1 | 0.75 | 0.66 |
| C5orf4 | 0.75 | 0.83 |
| TJP2 | 0.74 | 0.76 |
| KANK1 | 0.74 | 0.70 |
| APOLD1 | 0.74 | 0.58 |
| GPRC5B | 0.73 | 0.75 |
| TPCN1 | 0.73 | 0.71 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MED19 | -0.52 | -0.63 |
| NKIRAS2 | -0.50 | -0.53 |
| NR2C2AP | -0.49 | -0.61 |
| LSM7 | -0.49 | -0.68 |
| DUSP12 | -0.49 | -0.57 |
| PODXL2 | -0.49 | -0.54 |
| ZNF821 | -0.48 | -0.54 |
| TCTEX1D2 | -0.48 | -0.63 |
| POLB | -0.48 | -0.60 |
| CCDC28B | -0.48 | -0.66 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | TAS | 1621094 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007411 | axon guidance | TAS | axon (GO term level: 13) | 1621094 |
| GO:0007155 | cell adhesion | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 1621094 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| CORRE MULTIPLE MYELOMA UP | 74 | 45 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS UP | 17 | 11 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS DN | 133 | 77 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| CHEBOTAEV GR TARGETS DN | 120 | 73 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER B | 48 | 22 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL | 254 | 164 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| NIELSEN SCHWANNOMA UP | 18 | 16 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER HCP WITH H3K27ME3 | 97 | 72 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS UP | 170 | 107 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER DN | 74 | 34 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 UP | 141 | 75 | All SZGR 2.0 genes in this pathway |
| BOYAULT LIVER CANCER SUBCLASS G1 UP | 113 | 70 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 DN | 448 | 282 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 1479 | 1485 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-137 | 2432 | 2438 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| miR-140 | 1116 | 1122 | 1A | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-183 | 683 | 689 | 1A | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-214 | 249 | 255 | m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-218 | 263 | 269 | m8 | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-23 | 1497 | 1504 | 1A,m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-30-5p | 1575 | 1582 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-323 | 1497 | 1503 | 1A | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-330 | 771 | 777 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-410 | 2238 | 2244 | m8 | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-488 | 547 | 553 | m8 | hsa-miR-488 | CCCAGAUAAUGGCACUCUCAA |
| miR-493-5p | 1716 | 1722 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-495 | 2235 | 2241 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-496 | 448 | 454 | 1A | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-542-3p | 1525 | 1531 | 1A | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
| miR-543 | 1447 | 1453 | m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.