Gene Page: RIMS2
Summary ?
| GeneID | 9699 |
| Symbol | RIMS2 |
| Synonyms | OBOE|RAB3IP3|RIM2 |
| Description | regulating synaptic membrane exocytosis 2 |
| Reference | MIM:606630|HGNC:HGNC:17283|Ensembl:ENSG00000176406|HPRD:09436| |
| Gene type | protein-coding |
| Map location | 8q22.3 |
| Pascal p-value | 0.078 |
| Sherlock p-value | 0.92 |
| Fetal beta | -1.679 |
| DMG | 2 (# studies) |
| eGene | Cerebellar Hemisphere Myers' cis & trans Meta |
| Support | EXOCYTOSIS G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_clathrin G2Cdb.human_Synaptosome |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09637757 | 8 | 104831516 | RIMS2 | 4.897E-4 | 0.601 | 0.047 | DMG:Wockner_2014 |
| cg02663265 | 8 | 104513186 | RIMS2 | 3.09E-9 | -0.014 | 2.1E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2511587 | chr8 | 105256284 | RIMS2 | 9699 | 0.09 | cis | ||
| rs2441816 | chr8 | 105262972 | RIMS2 | 9699 | 0.08 | cis | ||
| rs2511595 | chr8 | 105272092 | RIMS2 | 9699 | 0.01 | cis | ||
| rs2511606 | chr8 | 105295694 | RIMS2 | 9699 | 0.09 | cis | ||
| rs2514631 | chr8 | 105309102 | RIMS2 | 9699 | 0.02 | cis |
Section II. Transcriptome annotation
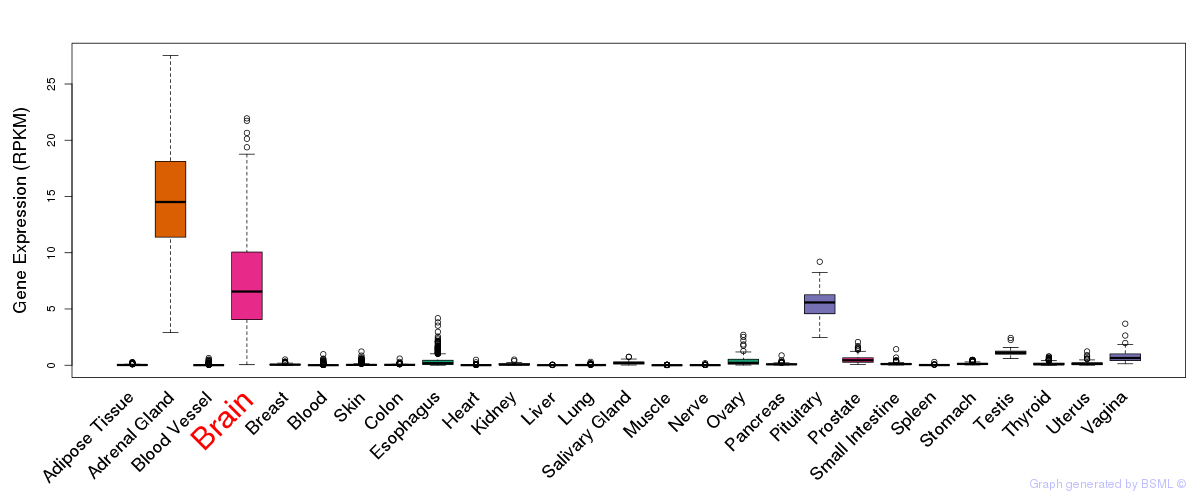
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AXL | 0.92 | 0.91 |
| ATP1A2 | 0.90 | 0.90 |
| SLC13A5 | 0.89 | 0.84 |
| S1PR1 | 0.89 | 0.90 |
| FAM167A | 0.89 | 0.86 |
| ACSBG1 | 0.88 | 0.84 |
| PLEKHO2 | 0.88 | 0.78 |
| ACSS1 | 0.88 | 0.90 |
| GLUL | 0.88 | 0.82 |
| MERTK | 0.87 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MED19 | -0.55 | -0.63 |
| CAMKV | -0.51 | -0.53 |
| PPAPR5 | -0.51 | -0.57 |
| ARL4D | -0.50 | -0.56 |
| LRRC7 | -0.50 | -0.54 |
| DPF1 | -0.50 | -0.57 |
| NOL4 | -0.49 | -0.53 |
| DUSP12 | -0.49 | -0.58 |
| PPP3CC | -0.49 | -0.56 |
| NELL2 | -0.49 | -0.55 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC3 TARGETS DN | 536 | 332 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 12HR DN | 209 | 122 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION UP | 552 | 347 | All SZGR 2.0 genes in this pathway |
| DACOSTA ERCC3 ALLELE XPCS VS TTD UP | 28 | 19 | All SZGR 2.0 genes in this pathway |
| TOMLINS METASTASIS UP | 14 | 9 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q12 Q22 AMPLICON | 132 | 82 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY MUTATED AND AMPLIFIED IN BREAST CANCER | 94 | 60 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| SUZUKI RESPONSE TO TSA AND DECITABINE 1B | 23 | 14 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| NIELSEN LEIOMYOSARCOMA CNN1 UP | 19 | 13 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| WANG LSD1 TARGETS DN | 39 | 30 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE DN | 23 | 15 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |
| KUMAR AUTOPHAGY NETWORK | 71 | 46 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |