Gene Page: SETD1A
Summary ?
| GeneID | 9739 |
| Symbol | SETD1A |
| Synonyms | KMT2F|Set1|Set1A |
| Description | SET domain containing 1A |
| Reference | MIM:611052|HGNC:HGNC:29010|Ensembl:ENSG00000099381|HPRD:13795|Vega:OTTHUMG00000150474 |
| Gene type | protein-coding |
| Map location | 16p11.2 |
| Pascal p-value | 0.515 |
| Sherlock p-value | 0.257 |
| Fetal beta | -0.273 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
| Support | Chromatin Remodeling Genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| DNM:Guipponi_2014 | Whole Exome Sequencing analysis | 49 DNMs were identified by comparing the exome of 53 individuals with sporadic SCZ and of their non-affected parents | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.01775 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SETD1A | chr16 | 30970148 | G | T | NM_014712 | . | silent | Schizophrenia | DNM:Xu_2012 | ||
| SETD1A | NM_014712 | c.4582delAG.- | splice | NA | NA | Schizophrenia | DNM:Guipponi_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26233914 | 16 | 31366536 | SETD1A | 0.002 | 3.945 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4141780 | chr20 | 40297792 | SETD1A | 9739 | 0.14 | trans |
Section II. Transcriptome annotation
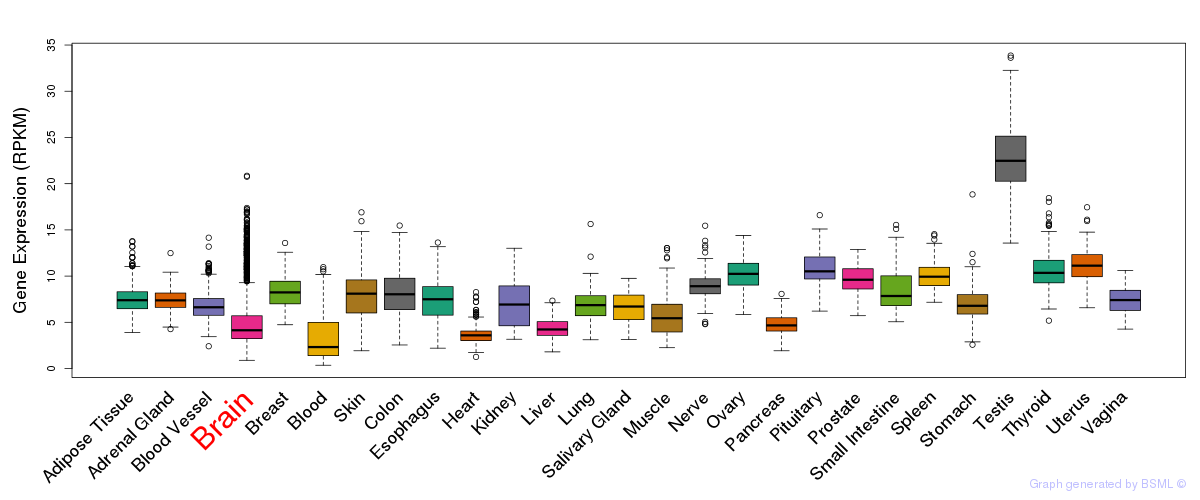
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| POM121C | 0.97 | 0.97 |
| SSH1 | 0.93 | 0.94 |
| UBN1 | 0.92 | 0.94 |
| GBF1 | 0.92 | 0.94 |
| TBC1D24 | 0.92 | 0.94 |
| RPAP1 | 0.92 | 0.94 |
| HCFC1 | 0.92 | 0.94 |
| PCNXL3 | 0.92 | 0.90 |
| ZFYVE20 | 0.92 | 0.92 |
| SEC16A | 0.91 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.69 | -0.73 |
| AF347015.21 | -0.68 | -0.81 |
| MT-CO2 | -0.68 | -0.73 |
| HIGD1B | -0.66 | -0.73 |
| C1orf54 | -0.66 | -0.83 |
| FXYD1 | -0.65 | -0.69 |
| GNG11 | -0.64 | -0.75 |
| MT-CYB | -0.64 | -0.67 |
| AF347015.8 | -0.64 | -0.70 |
| AF347015.33 | -0.64 | -0.66 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0003723 | RNA binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 12670868 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008168 | methyltransferase activity | IEA | - | |
| GO:0018024 | histone-lysine N-methyltransferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006350 | transcription | IEA | - | |
| GO:0016568 | chromatin modification | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0016607 | nuclear speck | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG LYSINE DEGRADATION | 44 | 29 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-142-5p | 626 | 632 | m8 | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-485-5p | 146 | 152 | 1A | hsa-miR-485-5p | AGAGGCUGGCCGUGAUGAAUUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.