Gene Page: MED12
Summary ?
| GeneID | 9968 |
| Symbol | MED12 |
| Synonyms | ARC240|CAGH45|FGS1|HOPA|MED12S|OHDOX|OKS|OPA1|TNRC11|TRAP230 |
| Description | mediator complex subunit 12 |
| Reference | MIM:300188|HGNC:HGNC:11957|Ensembl:ENSG00000184634|HPRD:02176|Vega:OTTHUMG00000021788 |
| Gene type | protein-coding |
| Map location | Xq13 |
| Sherlock p-value | 0.953 |
| DEG p-value | DEG:Zhao_2015:p=3.07e-04:q=0.0924 |
| Fetal beta | 0.162 |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Zhao_2015 | RNA Sequencing analysis | Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
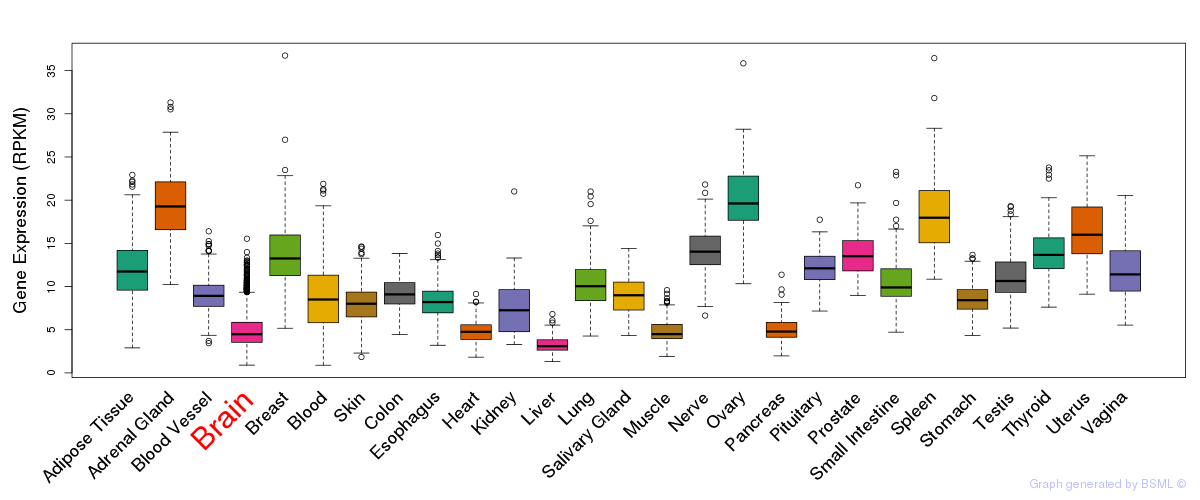
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ERAL1 | 0.88 | 0.85 |
| MCRS1 | 0.87 | 0.84 |
| TBRG4 | 0.87 | 0.85 |
| C11orf49 | 0.86 | 0.83 |
| SLC35B1 | 0.86 | 0.82 |
| JAGN1 | 0.86 | 0.83 |
| TMEM203 | 0.85 | 0.85 |
| ZDHHC16 | 0.85 | 0.82 |
| APEH | 0.85 | 0.83 |
| C7orf20 | 0.85 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.8 | -0.68 | -0.62 |
| AF347015.18 | -0.65 | -0.68 |
| MT-CO2 | -0.65 | -0.58 |
| AF347015.26 | -0.65 | -0.62 |
| AF347015.33 | -0.63 | -0.58 |
| MT-CYB | -0.63 | -0.59 |
| AF347015.31 | -0.63 | -0.58 |
| AF347015.27 | -0.63 | -0.60 |
| MT-ATP8 | -0.63 | -0.66 |
| AF347015.21 | -0.63 | -0.58 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IDA | 12218053 | |
| GO:0005515 | protein binding | IPI | 16109376 | |
| GO:0019904 | protein domain specific binding | IPI | 11984006 | |
| GO:0016455 | RNA polymerase II transcription mediator activity | IDA | 10198638 | |
| GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity | NAS | 10235266 | |
| GO:0042809 | vitamin D receptor binding | NAS | 10235266 | |
| GO:0046966 | thyroid hormone receptor binding | IDA | 10198638 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | IDA | 12218053 | |
| GO:0006350 | transcription | IEA | - | |
| GO:0030521 | androgen receptor signaling pathway | IDA | 12218053 | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IDA | 12037571 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000119 | mediator complex | IDA | 10198638 | |
| GO:0005813 | centrosome | IDA | 18029348 | |
| GO:0005634 | nucleus | IDA | 10235267 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005730 | nucleolus | IDA | 18029348 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CDK8 | K35 | MGC126074 | MGC126075 | cyclin-dependent kinase 8 | Affinity Capture-MS | BioGRID | 10198638 |
| LYST | CHS | CHS1 | lysosomal trafficking regulator | - | HPRD,BioGRID | 11984006 |
| MED10 | L6 | MGC5309 | NUT2 | TRG20 | mediator complex subunit 10 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 15175163 |
| MED19 | DT2P1G7 | LCMR1 | mediator complex subunit 19 | Affinity Capture-MS | BioGRID | 15175163 |
| MED26 | CRSP7 | CRSP70 | mediator complex subunit 26 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 15175163 |
| MED28 | 1500003D12Rik | DKFZp434N185 | EG1 | magicin | mediator complex subunit 28 | Affinity Capture-MS | BioGRID | 15175163 |
| MED29 | DKFZp434H247 | IXL | mediator complex subunit 29 | - | HPRD | 14576168 |
| MED29 | DKFZp434H247 | IXL | mediator complex subunit 29 | Affinity Capture-MS | BioGRID | 15175163 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | - | HPRD | 14638676 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | Affinity Capture-MS | BioGRID | 15175163 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | Reconstituted Complex | BioGRID | 12660246 |
| PPARGC1A | LEM6 | PGC-1(alpha) | PGC-1v | PGC1 | PGC1A | PPARGC1 | peroxisome proliferator-activated receptor gamma, coactivator 1 alpha | Affinity Capture-Western Reconstituted Complex | BioGRID | 14636573 |
| SOX9 | CMD1 | CMPD1 | SRA1 | SRY (sex determining region Y)-box 9 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12136106 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | Affinity Capture-MS Affinity Capture-Western | BioGRID | 10198638 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | Affinity Capture-MS | BioGRID | 10198638 |12837248 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| MAYBURD RESPONSE TO L663536 UP | 29 | 18 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS | 179 | 105 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| AMBROSINI FLAVOPIRIDOL TREATMENT TP53 | 109 | 63 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-23 | 143 | 149 | m8 | hsa-miR-23abrain | AUCACAUUGCCAGGGAUUUCC |
| hsa-miR-23bbrain | AUCACAUUGCCAGGGAUUACC | ||||
| miR-323 | 142 | 149 | 1A,m8 | hsa-miR-323brain | GCACAUUACACGGUCGACCUCU |
| miR-381 | 169 | 175 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-410 | 180 | 186 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-495 | 137 | 143 | 1A | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.