Gene Page: KCNE2
Summary ?
| GeneID | 9992 |
| Symbol | KCNE2 |
| Synonyms | ATFB4|LQT5|LQT6|MIRP1 |
| Description | potassium voltage-gated channel subfamily E regulatory subunit 2 |
| Reference | MIM:603796|HGNC:HGNC:6242|HPRD:04813| |
| Gene type | protein-coding |
| Map location | 21q22.12 |
| Pascal p-value | 0.767 |
| Fetal beta | 0.212 |
| eGene | Myers' cis & trans |
| Support | EXCITABILITY |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 2.507 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11701963 | chr21 | 35323656 | KCNE2 | 9992 | 0.18 | cis |
Section II. Transcriptome annotation
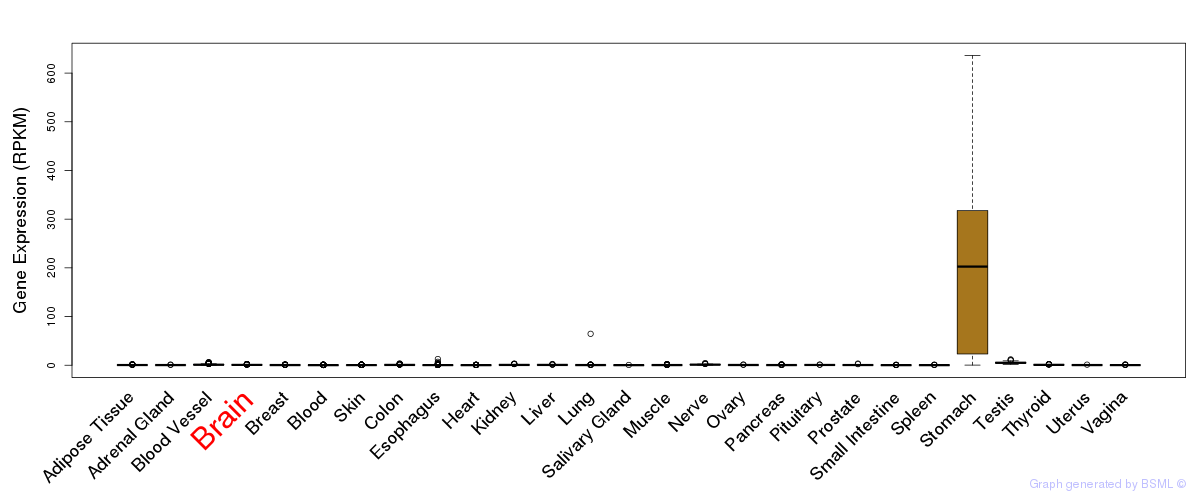
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| YTHDF2 | 0.97 | 0.96 |
| PAK2 | 0.96 | 0.96 |
| NUP160 | 0.96 | 0.92 |
| HSDL1 | 0.96 | 0.97 |
| SPAST | 0.96 | 0.97 |
| PBRM1 | 0.96 | 0.97 |
| ZNF776 | 0.96 | 0.96 |
| ZNF606 | 0.96 | 0.93 |
| PRRC1 | 0.96 | 0.96 |
| MIER3 | 0.96 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.74 | -0.81 |
| AIFM3 | -0.73 | -0.80 |
| C5orf53 | -0.73 | -0.78 |
| FXYD1 | -0.72 | -0.91 |
| MT-CO2 | -0.72 | -0.91 |
| S100B | -0.72 | -0.85 |
| AF347015.31 | -0.71 | -0.88 |
| AF347015.33 | -0.71 | -0.87 |
| TSC22D4 | -0.71 | -0.81 |
| AF347015.27 | -0.70 | -0.86 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0005249 | voltage-gated potassium channel activity | TAS | 10219239 | |
| GO:0030955 | potassium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008015 | blood circulation | NAS | 10219239 | |
| GO:0008016 | regulation of heart contraction | TAS | 10219239 | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006813 | potassium ion transport | TAS | 10219239 | |
| GO:0006936 | muscle contraction | TAS | 10219239 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005764 | lysosome | IDA | 16780588 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 16780588 | |
| GO:0008076 | voltage-gated potassium channel complex | TAS | 10219239 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| LEIN CHOROID PLEXUS MARKERS | 103 | 61 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |