| General information | Literature | Expression | Regulation | Variant | Interaction |
Basic Information | |
|---|---|
Gene ID | 2272 |
Name | FHIT |
Synonymous | fragile histidine triad;FHIT;fragile histidine triad |
Definition | AP3A hydrolase|bis(5'-adenosyl)-triphosphatase|diadenosine 5',5'''-P1,P3-triphosphate hydrolase|dinucleosidetriphosphatase|tumor suppressor protein |
Position | 3p14.2 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
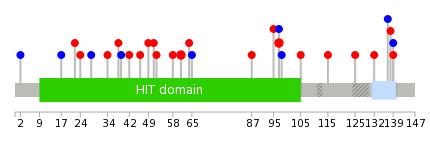 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
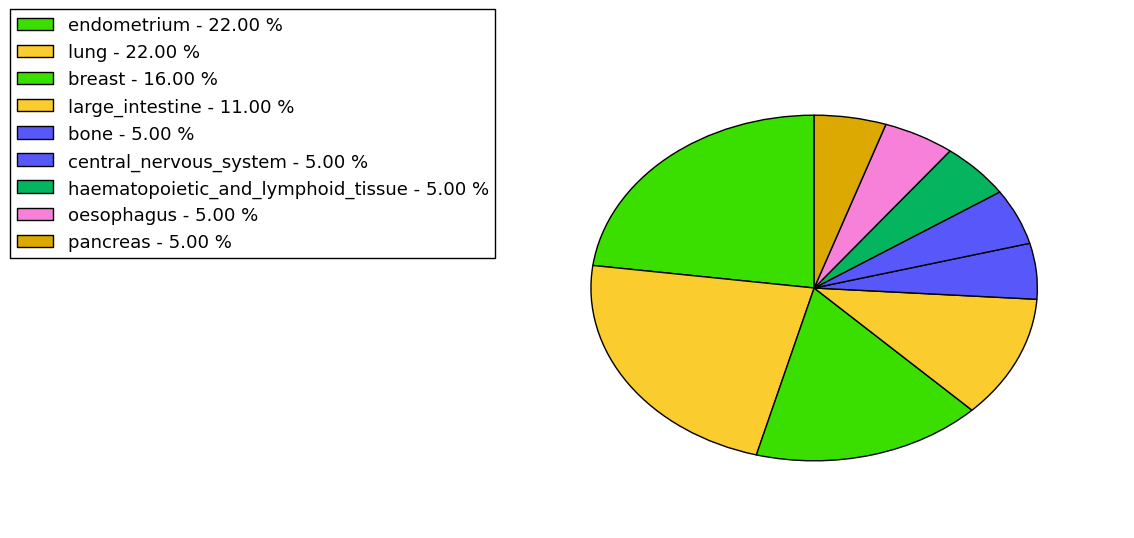 |
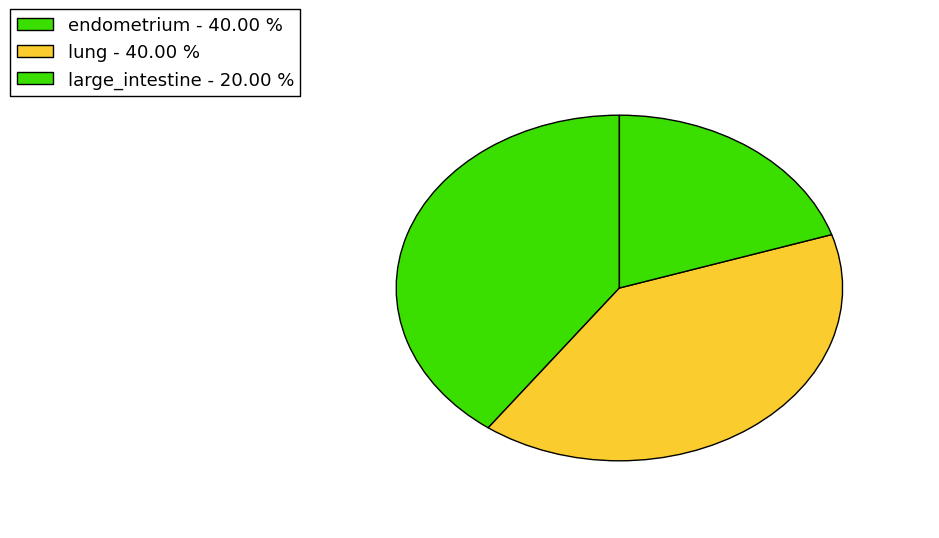 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.06. |
The copy number variations for various cancers | Top |
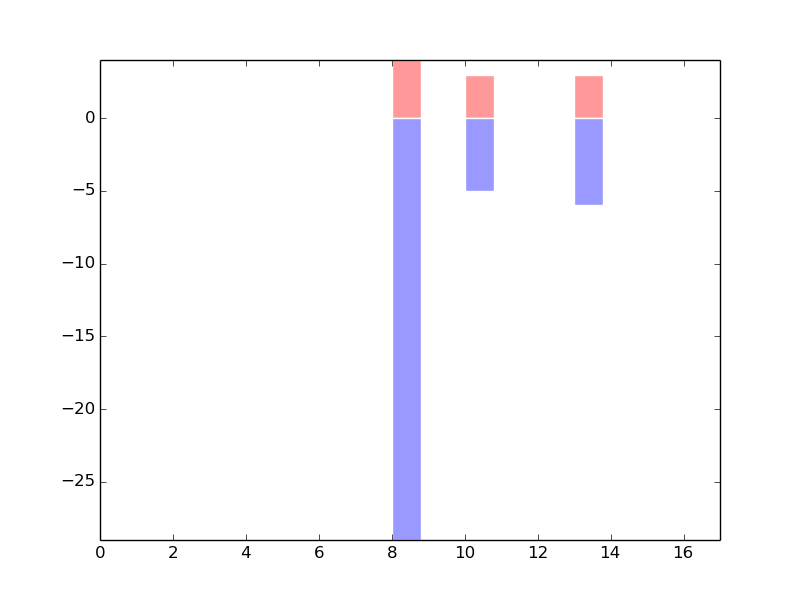 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
Mutation (CDS; AA; Chr) | Site | Histology | Mutation Type |
|---|---|---|---|
c.313G>C; p.G105R; 3:59922381-59922381 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.291C>T; p.V97V; 3:59922403-59922403 |
skin | malignant_melanoma | Substitution - coding silent |
c.289G>A; p.V97I; 3:59922405-59922405 |
oesophagus | carcinoma; adenocarcinoma | Substitution - Missense |
c.289G>A; p.V97I; 3:59922405-59922405 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.113T>C; p.V38A; 3:60014143-60014143 |
prostate | carcinoma; adenocarcinoma | Substitution - Missense |
c.411A>T; p.A137A; 3:59752259-59752259 |
lung | carcinoma; adenocarcinoma | Substitution - coding silent |
c.117C>T; p.C39C; 3:60014139-60014139 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.294T>C; p.H98H; 3:59922400-59922400 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.191G>C; p.R64T; 3:60014065-60014065 |
breast | carcinoma | Substitution - Missense |
c.417C>T; p.A139A; 3:59752253-59752253 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.84G>A; p.R28R; 3:60536879-60536879 |
skin | malignant_melanoma | Substitution - coding silent |
c.64T>C; p.S22P; 3:60536899-60536899 |
breast | carcinoma | Substitution - Missense |
c.259G>A; p.E87K; 3:60011391-60011391 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Missense |
c.414A>C; p.E138D; 3:59752256-59752256 |
skin | malignant_melanoma | Substitution - Missense |
c.345G>T; p.E115D; 3:59922349-59922349 |
pancreas | carcinoma | Substitution - Missense |
c.101G>T; p.G34V; 3:60536862-60536862 |
lung | carcinoma; small_cell_carcinoma | Substitution - Missense |
c.6G>C; p.S2S; 3:60536957-60536957 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.152G>A; p.R51H; 3:60014104-60014104 |
breast | carcinoma | Substitution - Missense |
c.182C>T; p.T61M; 3:60014074-60014074 |
stomach | adenocarcinoma | Substitution - Missense |
c.182C>T; p.T61M; 3:60014074-60014074 |
bone; pelvis | chondrosarcoma | Substitution - Missense |
c.137G>A; p.R46H; 3:60014119-60014119 |
central_nervous_system; brain | glioma; astrocytoma_Grade_IV | Substitution - Missense |
c.49C>A; p.L17I; 3:60536914-60536914 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.195C>T; p.V65V; 3:60014061-60014061 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.70G>A; p.A24T; 3:60536893-60536893 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.100G>T; p.G34*; 3:60536863-60536863 |
lung | carcinoma; small_cell_carcinoma | Substitution - Nonsense |
c.103C>A; p.H35N; 3:60536860-60536860 |
skin; neck | malignant_melanoma | Substitution - Missense |
c.263C>T; p.A88V; 3:60011387-60011387 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.263C>T; p.A88V; 3:60011387-60011387 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.124C>T; p.R42W; 3:60014132-60014132 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.283G>C; p.V95L; 3:59922411-59922411 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.349-9_349-8insT; p.?; 3:59752329-59752330 |
skin | malignant_melanoma | Unknown |
c.145G>T; p.D49Y; 3:60014111-60014111 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.155C>T; p.P52L; 3:60014101-60014101 |
skin | malignant_melanoma | Substitution - Missense |
c.416C>T; p.A139V; 3:59752254-59752254 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.370G>T; p.D124Y; 3:59752300-59752300 |
skin | malignant_melanoma | Substitution - Missense |
c.51C>G; p.L17L; 3:60536912-60536912 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.262G>A; p.A88T; 3:60011388-60011388 |
stomach | carcinoma; intestinal_adenocarcinoma | Substitution - Missense |
c.374T>A; p.F125Y; 3:59752296-59752296 |
skin | malignant_melanoma | Substitution - Missense |
c.174G>T; p.L58F; 3:60014082-60014082 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; plasma_cell_myeloma | Substitution - Missense |
c.394G>C; p.E132Q; 3:59752276-59752276 |
NS | NS | Substitution - Missense |