| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 26038 |
Name | CHD5 |
Synonymous | chromodomain helicase DNA binding protein 5;CHD5;chromodomain helicase DNA binding protein 5 |
Definition | ATP-dependent helicase CHD5|chromodomain-helicase-DNA-binding protein 5 |
Position | 1p36.31 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
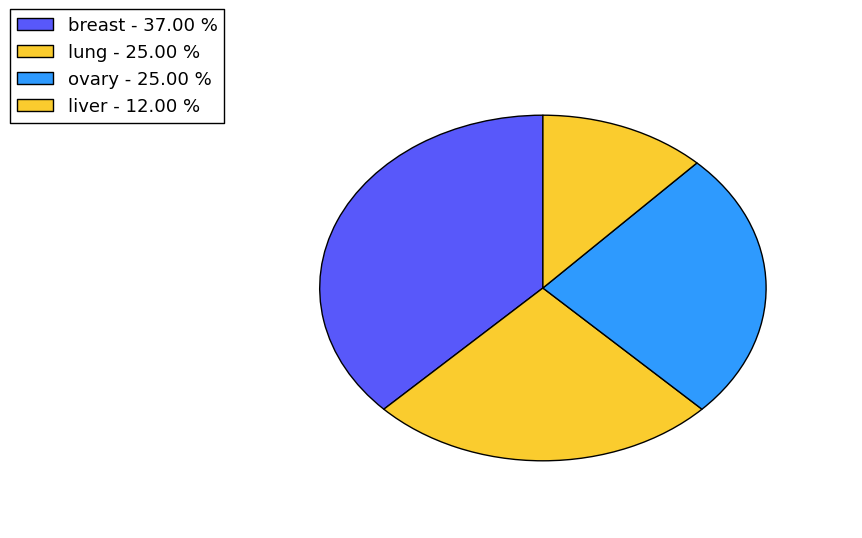 |
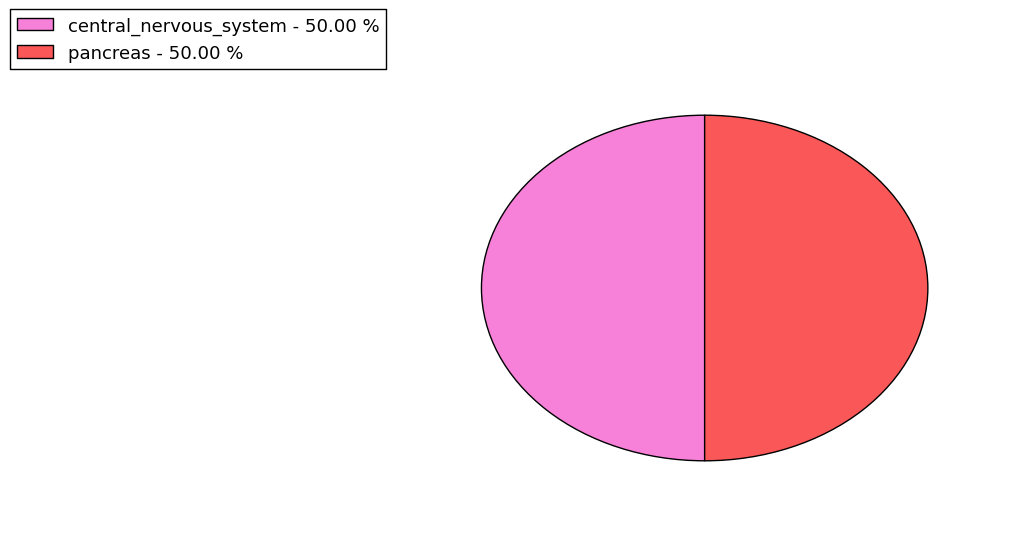 |
Loss of Function mutations compare to missense mutations | Top |
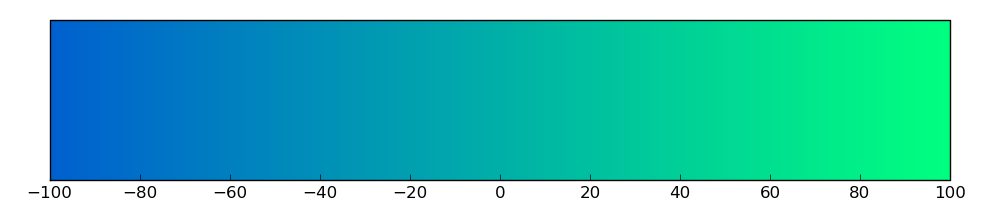 |
| The ratio of the loss-of-function mutations over missense mutations is 0.20. |
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.355G>A; p.D119N; 1:6159368-6159368 |
breast | carcinoma; ductal_carcinoma | Substitution - Missense |
c.1972A>G; p.R658G; 1:6143894-6143894 |
liver | carcinoma; hepatocellular_carcinoma | Substitution - Missense |
c.5109C>T; p.F1703F; 1:6112171-6112171 |
skin; arm | malignant_melanoma | Substitution - coding silent |
c.3969_3970insAT; p.L1324fs*78; 1:6126680-6126681 |
ovary | carcinoma; serous_carcinoma | Insertion - Frameshift |
c.3499A>G; p.M1167V; 1:6128958-6128958 |
ovary | carcinoma; serous_carcinoma | Substitution - Missense |
c.133G>A; p.V45M; 1:6168224-6168224 |
breast | carcinoma; ductal_carcinoma | Substitution - Missense |
c.5622C>T; p.D1874D; 1:6106736-6106736 |
pancreas | NS | Substitution - coding silent |
c.898G>C; p.D300H; 1:6151128-6151128 |
upper_aerodigestive_tract; mouth | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.1391_1392insC; p.L465fs*28; 1:6146863-6146864 |
ovary | carcinoma; serous_carcinoma | Insertion - Frameshift |
c.3917G>T; p.R1306L; 1:6126733-6126733 |
lung | carcinoma; small_cell_carcinoma | Substitution - Missense |
c.3917G>T; p.R1306L; 1:6126733-6126733 |
lung | carcinoma; small_cell_carcinoma | Substitution - Missense |
c.5199C>T; p.Y1733Y; 1:6111825-6111825 |
central_nervous_system; brain | glioma; astrocytoma_Grade_IV | Substitution - coding silent |
c.3772G>C; p.D1258H; 1:6128177-6128177 |
ovary | carcinoma; clear_cell_carcinoma | Substitution - Missense |
c.108C>T; p.F36F; 1:6168249-6168249 |
upper_aerodigestive_tract; pharynx | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.1999A>G; p.R667G; 1:6143867-6143867 |
breast | carcinoma; ductolobular_carcinoma | Substitution - Missense |