| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 3065 |
Name | HDAC1 |
Synonymous | histone deacetylase 1;HDAC1;histone deacetylase 1 |
Definition | reduced potassium dependency, yeast homolog-like 1 |
Position | 1p34 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
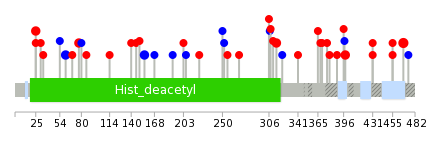 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
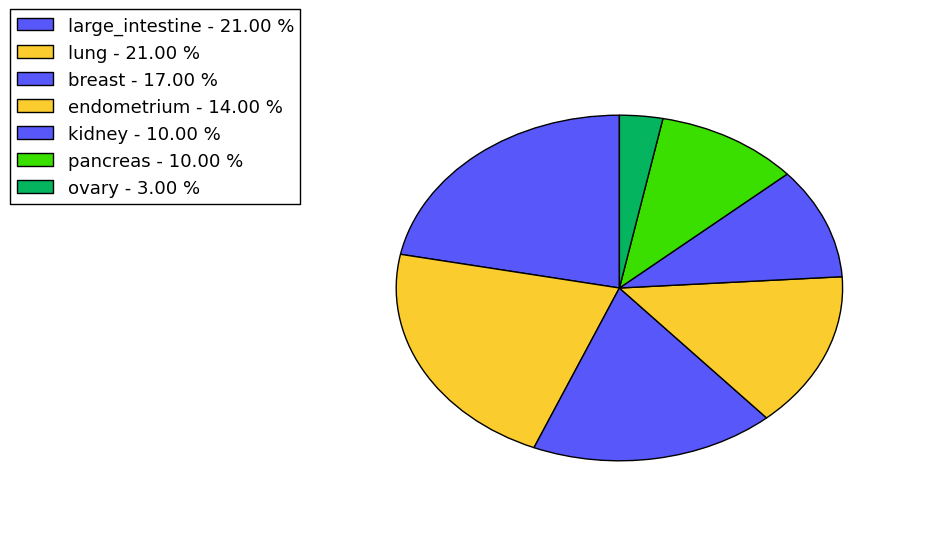 |
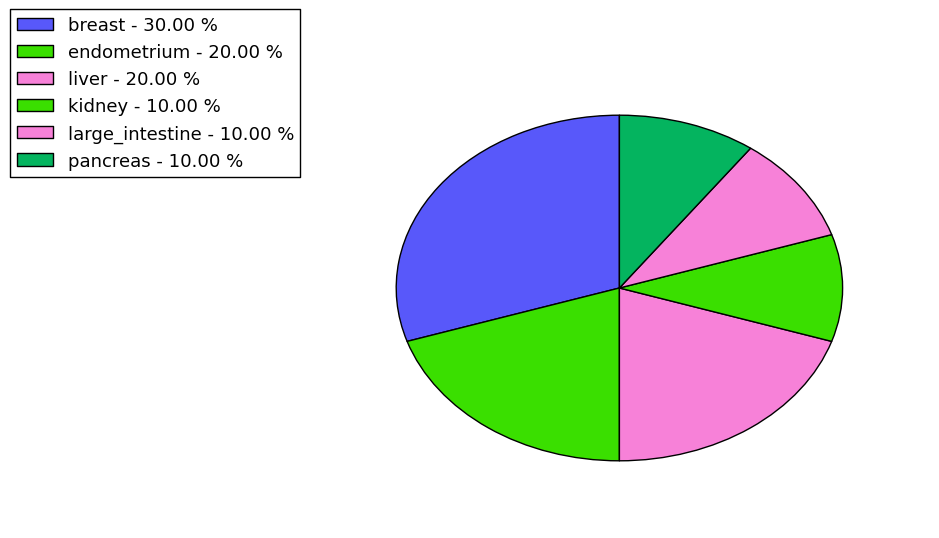 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.18. |
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.1191C>T; p.D397D; 1:32331778-32331778 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.943G>A; p.E315K; 1:32330872-32330872 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.92A>C; p.K31T; 1:32302663-32302663 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Missense |
c.943G>A; p.E315K; 1:32330872-32330872 |
urinary_tract; bladder | carcinoma; transitional_cell_carcinoma | Substitution - Missense |
c.74G>C; p.G25A; 1:32302645-32302645 |
ovary | carcinoma; serous_carcinoma | Substitution - Missense |
c.767C>T; p.A256V; 1:32330615-32330615 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.1397_1399delAGG; p.E468delE; 1:32332725-32332727 |
large_intestine; colon | carcinoma; adenocarcinoma | Deletion - In frame |
c.1397_1399delAGG; p.E468delE; 1:32332725-32332727 |
large_intestine; colon | carcinoma; adenocarcinoma | Deletion - In frame |
c.1397_1399delAGG; p.E468delE; 1:32332725-32332727 |
large_intestine | carcinoma; adenocarcinoma | Deletion - In frame |
c.1397_1399delAGG; p.E468delE; 1:32332725-32332727 |
large_intestine | carcinoma; adenocarcinoma | Deletion - In frame |
c.795C>T; p.S265S; 1:32330643-32330643 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.1397_1399delAGG; p.E468delE; 1:32332725-32332727 |
endometrium | carcinoma; endometrioid_carcinoma | Deletion - In frame |
c.436G>A; p.E146K; 1:32327019-32327019 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.449T>G; p.F150C; 1:32327032-32327032 |
pancreas | carcinoma | Substitution - Missense |
c.540T>C; p.G180G; 1:32327581-32327581 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.1108C>T; p.L370F; 1:32331695-32331695 |
skin; scalp | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.183T>A; p.A61A; 1:32316685-32316685 |
liver | carcinoma | Substitution - coding silent |
c.183T>A; p.A61A; 1:32316685-32316685 |
liver | carcinoma | Substitution - coding silent |
c.1023C>G; p.F341L; 1:32331517-32331517 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.229C>T; p.R77C; 1:32316731-32316731 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.229C>T; p.R77C; 1:32316731-32316731 |
lung | carcinoma; bronchioloalveolar_adenocarcinoma | Substitution - Missense |
c.1335_1336delAG; p.E446fs*2; 1:32332205-32332206 |
breast | carcinoma | Deletion - Frameshift |
c.1162G>A; p.A388T; 1:32331749-32331749 |
lung | carcinoid-endocrine_tumour; atypical | Substitution - Missense |
c.1162G>A; p.A388T; 1:32331749-32331749 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.921C>T; p.N307N; 1:32330850-32330850 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - coding silent |
c.286G>A; p.V96I; 1:32324484-32324484 |
breast | carcinoma | Substitution - Missense |
c.1422G>A; p.G474G; 1:32333017-32333017 |
breast | carcinoma | Substitution - coding silent |
c.844G>A; p.A282T; 1:32330773-32330773 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.1422-2A>T; p.?; 1:32333015-32333015 |
breast | carcinoma | Unknown |
c.1103A>G; p.E368G; 1:32331690-32331690 |
skin | malignant_melanoma | Substitution - Missense |
c.685C>T; p.R229*; 1:32329116-32329116 |
stomach | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.74G>A; p.G25E; 1:32302645-32302645 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.74G>A; p.G25E; 1:32302645-32302645 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.232T>C; p.S78P; 1:32316734-32316734 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.1188G>C; p.E396D; 1:32331775-32331775 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.258G>C; p.E86D; 1:32316760-32316760 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.418C>T; p.H140Y; 1:32327001-32327001 |
breast | carcinoma; basal_(triple-negative)_carcinoma | Substitution - Missense |
c.618A>G; p.P206P; 1:32327659-32327659 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.895G>T; p.G299*; 1:32330824-32330824 |
lung | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.1090C>T; p.Q364*; 1:32331677-32331677 |
urinary_tract; bladder | carcinoma | Substitution - Nonsense |
c.839-2_839-1insG; p.?; 1:32330766-32330767 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Unknown |
c.1093C>T; p.R365*; 1:32331680-32331680 |
thyroid | carcinoma; anaplastic_carcinoma | Substitution - Nonsense |
c.1372+9G>A; p.?; 1:32332251-32332251 |
pancreas | carcinoma | Unknown |
c.750G>A; p.E250E; 1:32330598-32330598 |
skin | malignant_melanoma | Substitution - coding silent |
c.49+1G>C; p.?; 1:32292219-32292219 |
liver | carcinoma | Unknown |
c.637-1G>A; p.?; 1:32329067-32329067 |
skin | malignant_melanoma | Unknown |
c.809G>A; p.R270Q; 1:32330657-32330657 |
pancreas | carcinoma | Substitution - Missense |
c.1192G>A; p.E398K; 1:32331779-32331779 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.1192G>A; p.E398K; 1:32331779-32331779 |
breast | carcinoma | Substitution - Missense |
c.50-2A>G; p.?; 1:32302619-32302619 |
breast | carcinoma | Unknown |
c.609G>T; p.E203D; 1:32327650-32327650 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.1363G>A; p.E455K; 1:32332233-32332233 |
skin; trunk | malignant_melanoma | Substitution - Missense |
c.966G>T; p.T322T; 1:32330895-32330895 |
breast | carcinoma | Substitution - coding silent |
c.839-2A>G; p.?; 1:32330766-32330766 |
liver | carcinoma | Unknown |
c.237C>T; p.I79I; 1:32316739-32316739 |
skin; head_neck | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.281-2A>T; p.?; 1:32324477-32324477 |
oesophagus | carcinoma; adenocarcinoma | Unknown |
c.206G>A; p.S69N; 1:32316708-32316708 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.199T>C; p.Y67H; 1:32316701-32316701 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.199T>C; p.Y67H; 1:32316701-32316701 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.570G>A; p.T190T; 1:32327611-32327611 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.468C>T; p.I156I; 1:32327051-32327051 |
skin | malignant_melanoma | Substitution - coding silent |
c.468C>T; p.I156I; 1:32327051-32327051 |
NS | NS | Substitution - coding silent |
c.162T>C; p.Y54Y; 1:32302733-32302733 |
pancreas | carcinoma | Substitution - coding silent |
c.691G>A; p.G231R; 1:32329122-32329122 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.1135G>T; p.V379F; 1:32331722-32331722 |
breast | carcinoma | Substitution - Missense |
c.932G>A; p.C311Y; 1:32330861-32330861 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.1291C>T; p.R431C; 1:32332161-32332161 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.1292G>A; p.R431H; 1:32332162-32332162 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.1264G>A; p.D422N; 1:32332134-32332134 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.101G>A; p.R34Q; 1:32302672-32302672 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.101G>A; p.R34Q; 1:32302672-32302672 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.1094G>A; p.R365Q; 1:32331681-32331681 |
breast | carcinoma; basal_(triple-negative)_carcinoma | Substitution - Missense |
c.504G>A; p.Q168Q; 1:32327545-32327545 |
breast | carcinoma | Substitution - coding silent |
c.1126G>A; p.A376T; 1:32331713-32331713 |
breast | carcinoma | Substitution - Missense |
c.922G>A; p.V308I; 1:32330851-32330851 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.240T>C; p.R80R; 1:32316742-32316742 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - coding silent |
c.917G>T; p.R306L; 1:32330846-32330846 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.756C>T; p.F252F; 1:32330604-32330604 |
skin | malignant_melanoma | Substitution - coding silent |
c.341C>T; p.T114I; 1:32324539-32324539 |
pancreas | carcinoma | Substitution - Missense |
c.664T>G; p.Y222D; 1:32329095-32329095 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Missense |
c.813A>G; p.L271L; 1:32330661-32330661 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.1360_1362delGAG; p.E455delE; 1:32332230-32332232 |
endometrium | carcinoma; endometrioid_carcinoma | Deletion - In frame |
c.1381G>A; p.E461K; 1:32332709-32332709 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |