| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 7832 |
Name | BTG2 |
Synonymous | BTG family, member 2;BTG2;BTG family, member 2 |
Definition | B-cell translocation gene 2|NGF-inducible anti-proliferative protein PC3|nerve growth factor-inducible anti-proliferative|pheochromacytoma cell-3|protein BTG2 |
Position | 1q32 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
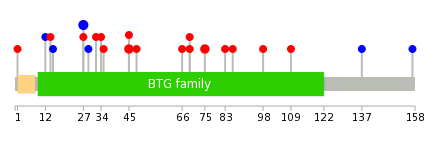 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
 |
 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.06. |
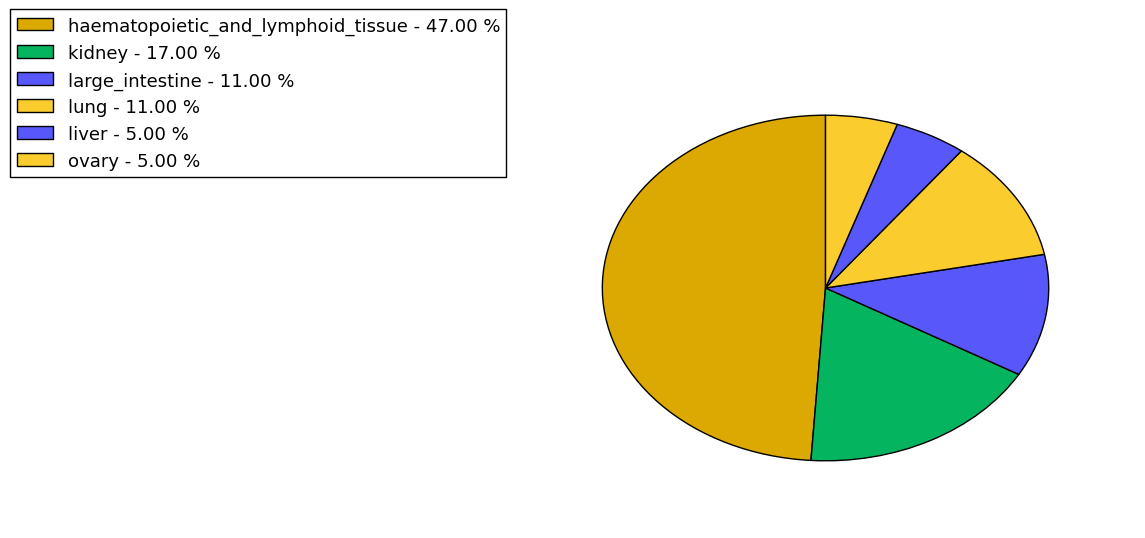
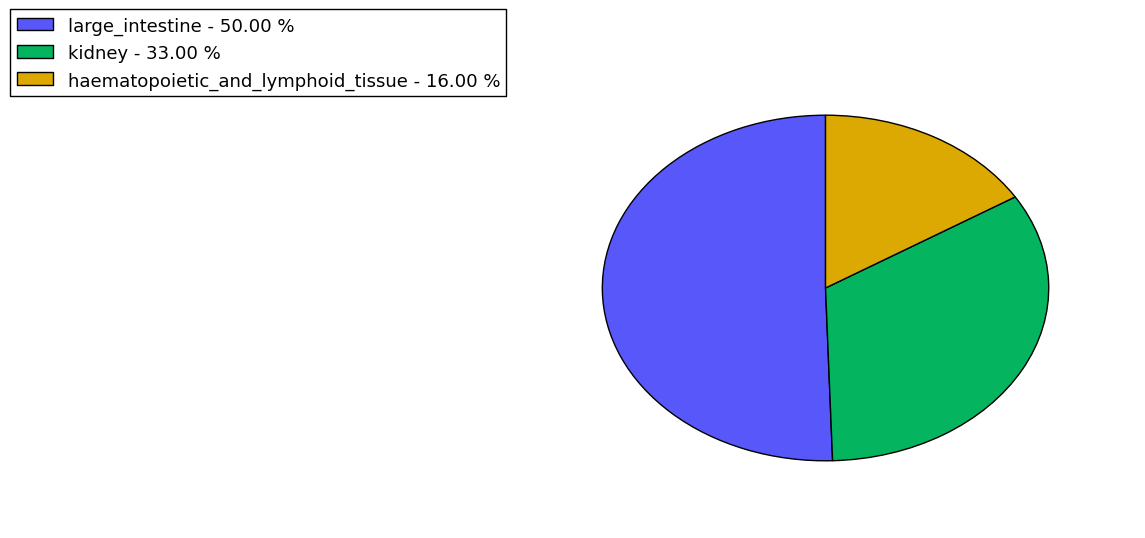
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.325G>A; p.V109M; 1:203307286-203307286 |
liver | carcinoma | Substitution - Missense |
c.87C>T; p.C29C; 1:203305693-203305693 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.133G>A; p.A45T; 1:203305739-203305739 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; diffuse_large_B_cell_lymphoma | Substitution - Missense |
c.133G>A; p.A45T; 1:203305739-203305739 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm | Substitution - Missense |
c.415A>G; p.N139D; 1:203307376-203307376 |
soft_tissue; striated_muscle | rhabdomyosarcoma; alveolar | Substitution - Missense |
c.134C>A; p.A45E; 1:203305740-203305740 |
haematopoietic_and_lymphoid_tissue; lymph_node | lymphoid_neoplasm; diffuse_large_B_cell_lymphoma | Substitution - Missense |
c.205C>G; p.R69G; 1:203307166-203307166 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.36G>A; p.E12E; 1:203305642-203305642 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; chronic_lymphocytic_leukaemia-small_lymphocytic_lymphoma | Substitution - coding silent |
c.103C>T; p.L35F; 1:203305709-203305709 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm | Substitution - Missense |
c.74G>A; p.R25K; 1:203305680-203305680 |
haematopoietic_and_lymphoid_tissue; lymph_node | lymphoid_neoplasm; follicular_lymphoma | Substitution - Missense |
c.396T>A; p.C132*; 1:203307357-203307357 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Nonsense |
c.81G>A; p.R27R; 1:203305687-203305687 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.81G>A; p.R27R; 1:203305687-203305687 |
skin | malignant_melanoma | Substitution - coding silent |
c.81G>A; p.R27R; 1:203305687-203305687 |
skin | malignant_melanoma | Substitution - coding silent |
c.96G>T; p.E32D; 1:203305702-203305702 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm | Substitution - Missense |
c.471C>T; p.S157S; 1:203307432-203307432 |
kidney | carcinoma; papillary_renal_cell_carcinoma | Substitution - coding silent |
c.41C>T; p.A14V; 1:203305647-203305647 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm | Substitution - Missense |
c.272A>G; p.Q91R; 1:203307233-203307233 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.411C>T; p.C137C; 1:203307372-203307372 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - coding silent |
c.293G>C; p.S98T; 1:203307254-203307254 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.45C>T; p.A15A; 1:203305651-203305651 |
kidney | carcinoma; papillary_renal_cell_carcinoma | Substitution - coding silent |
c.205C>T; p.R69C; 1:203307166-203307166 |
skin | malignant_melanoma | Substitution - Missense |
c.3G>A; p.M1I; 1:203305609-203305609 |
ovary | carcinoma; serous_carcinoma | Substitution - Missense |
c.102G>T; p.R34S; 1:203305708-203305708 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm | Substitution - Missense |
c.142+5G>A; p.?; 1:203305753-203305753 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm | Unknown |
c.257G>A; p.G86E; 1:203307218-203307218 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.223G>A; p.D75N; 1:203307184-203307184 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.223G>A; p.D75N; 1:203307184-203307184 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.196C>G; p.R66G; 1:203307157-203307157 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.249C>G; p.S83R; 1:203307210-203307210 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.80G>A; p.R27Q; 1:203305686-203305686 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.157C>G; p.H53D; 1:203307118-203307118 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.142G>C; p.E48Q; 1:203305748-203305748 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm | Substitution - Missense |