| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 9076 |
Name | CLDN1 |
Synonymous | claudin 1;CLDN1;claudin 1 |
Definition | claudin-1|senescence-associated epithelial membrane protein 1 |
Position | 3q28-q29 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
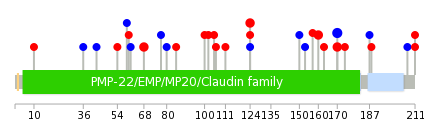 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
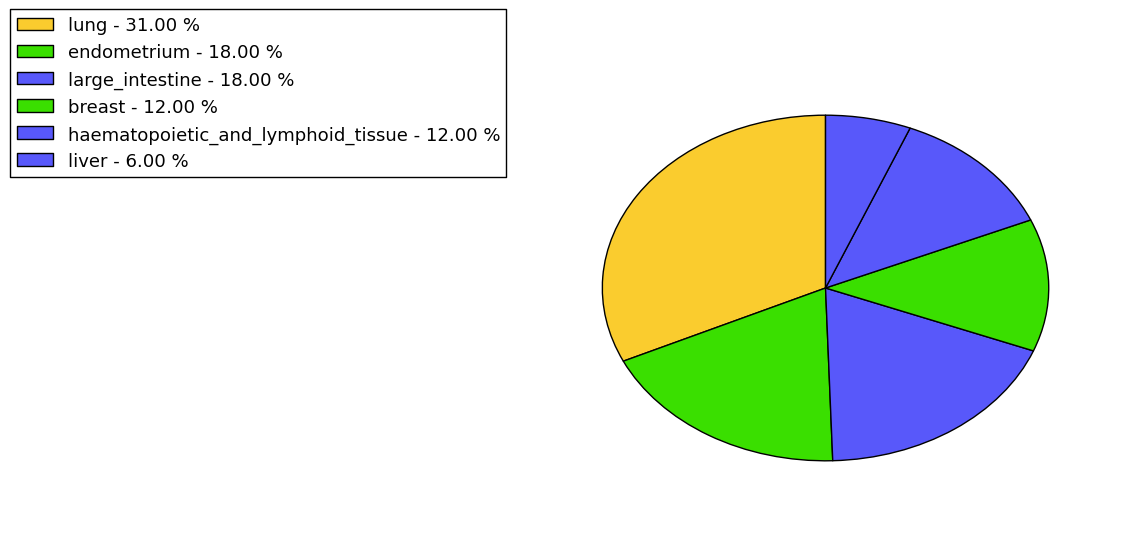 |
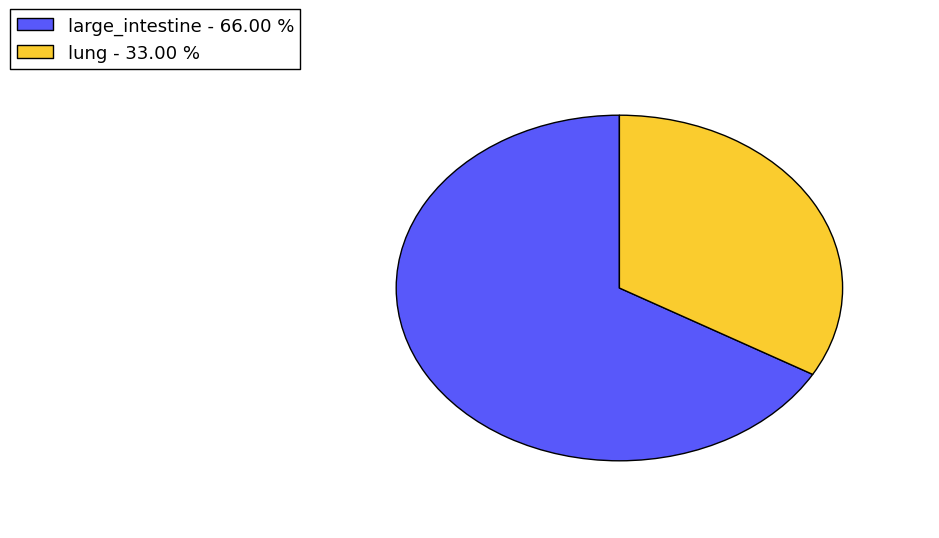 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.18. |
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.108C>T; p.A36A; 3:190322099-190322099 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.459C>T; p.T153T; 3:190310183-190310183 |
skin | malignant_melanoma | Substitution - coding silent |
c.181C>T; p.Q61*; 3:190322026-190322026 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Nonsense |
c.634T>C; p.*212R; 3:190308279-190308279 |
liver | carcinoma | Nonstop extension |
c.177C>T; p.T59T; 3:190322030-190322030 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - coding silent |
c.470C>T; p.A157V; 3:190310172-190310172 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.169C>T; p.Q57*; 3:190322038-190322038 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.202G>A; p.D68N; 3:190322005-190322005 |
urinary_tract; bladder | carcinoma; transitional_cell_carcinoma | Substitution - Missense |
c.202G>A; p.D68N; 3:190322005-190322005 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.439G>T; p.E147*; 3:190310203-190310203 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.439G>T; p.E147*; 3:190310203-190310203 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.121G>A; p.V41M; 3:190322086-190322086 |
oesophagus; lower_third | carcinoma; adenocarcinoma | Substitution - Missense |
c.631G>C; p.V211L; 3:190308282-190308282 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.636A>C; p.*212C; 3:190308277-190308277 |
lung | carcinoma; squamous_cell_carcinoma | Nonstop extension |
c.370G>A; p.A124T; 3:190312890-190312890 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.129C>T; p.A43A; 3:190322078-190322078 |
skin | malignant_melanoma | Substitution - coding silent |
c.371C>T; p.A124V; 3:190312889-190312889 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; chronic_lymphocytic_leukaemia-small_lymphocytic_lymphoma | Substitution - Missense |
c.370G>A; p.A124T; 3:190312890-190312890 |
lung; right_upper_lobe | carcinoma; adenocarcinoma | Substitution - Missense |
c.562C>G; p.R188G; 3:190308351-190308351 |
liver | carcinoma | Substitution - Missense |
c.298G>A; p.V100I; 3:190312962-190312962 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.600A>C; p.P200P; 3:190308313-190308313 |
oesophagus | carcinoma; adenocarcinoma | Substitution - coding silent |
c.142G>A; p.E48K; 3:190322065-190322065 |
stomach | carcinoma; diffuse_adenocarcinoma | Substitution - Missense |
c.461C>T; p.P154L; 3:190310181-190310181 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.331G>A; p.D111N; 3:190312929-190312929 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.331G>A; p.D111N; 3:190312929-190312929 |
skin | malignant_melanoma | Substitution - Missense |
c.509C>T; p.A170V; 3:190308404-190308404 |
lung | carcinoma; small_cell_carcinoma | Substitution - Missense |
c.160T>C; p.C54R; 3:190322047-190322047 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.509C>T; p.A170V; 3:190308404-190308404 |
lung | carcinoma; small_cell_carcinoma | Substitution - Missense |
c.183G>A; p.Q61Q; 3:190322024-190322024 |
skin | malignant_melanoma | Substitution - coding silent |
c.254T>G; p.V85G; 3:190313006-190313006 |
breast | carcinoma | Substitution - Missense |
c.631G>A; p.V211M; 3:190308282-190308282 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.561C>T; p.P187P; 3:190308352-190308352 |
skin | malignant_melanoma | Substitution - coding silent |
c.57C>T; p.I19I; 3:190322150-190322150 |
biliary_tract; gallbladder | carcinoma; adenocarcinoma | Substitution - coding silent |
c.60C>T; p.G20G; 3:190322147-190322147 |
large_intestine | carcinoma; adenocarcinoma | Substitution - coding silent |
c.621G>A; p.G207G; 3:190308292-190308292 |
skin | malignant_melanoma | Substitution - coding silent |
c.194_195delAA; p.K65fs*3; 3:190322012-190322013 |
large_intestine; colon | carcinoma; adenocarcinoma | Deletion - Frameshift |
c.473+5C>T; p.?; 3:190310164-190310164 |
oesophagus; lower_third | carcinoma; adenocarcinoma | Unknown |
c.372G>A; p.A124A; 3:190312888-190312888 |
skin | malignant_melanoma | Substitution - coding silent |
c.510T>C; p.A170A; 3:190308403-190308403 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.510T>C; p.A170A; 3:190308403-190308403 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.510T>C; p.A170A; 3:190308403-190308403 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.428G>T; p.R143I; 3:190310214-190310214 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.314T>C; p.M105T; 3:190312946-190312946 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.478G>A; p.E160K; 3:190308435-190308435 |
skin | malignant_melanoma | Substitution - Missense |
c.478G>A; p.E160K; 3:190308435-190308435 |
skin | malignant_melanoma | Substitution - Missense |
c.488A>T; p.Q163L; 3:190308425-190308425 |
breast | carcinoma | Substitution - Missense |
c.231G>A; p.L77L; 3:190313029-190313029 |
stomach | carcinoma; adenocarcinoma | Substitution - coding silent |
c.520C>T; p.L174F; 3:190308393-190308393 |
skin | malignant_melanoma | Substitution - Missense |
c.306G>A; p.M102I; 3:190312954-190312954 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; diffuse_large_B_cell_lymphoma | Substitution - Missense |
c.240C>T; p.T80T; 3:190313020-190313020 |
skin | malignant_melanoma | Substitution - coding silent |
c.178G>A; p.G60R; 3:190322029-190322029 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.325G>C; p.E109Q; 3:190312935-190312935 |
haematopoietic_and_lymphoid_tissue; soft_tissue | lymphoid_neoplasm; diffuse_large_B_cell_lymphoma | Substitution - Missense |
c.28G>C; p.G10R; 3:190322179-190322179 |
lung | carcinoma; squamous_cell_carcinoma | Substitution - Missense |
c.336G>T; p.E112D; 3:190312924-190312924 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.336G>T; p.E112D; 3:190312924-190312924 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.450C>T; p.D150D; 3:190310192-190310192 |
lung | carcinoma; adenocarcinoma | Substitution - coding silent |
c.316A>G; p.K106E; 3:190312944-190312944 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |