| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 94241 |
Name | TP53INP1 |
Synonymous | tumor protein p53 inducible nuclear protein 1;TP53INP1;tumor protein p53 inducible nuclear protein 1 |
Definition | p53-dependent damage-inducible nuclear protein 1|p53-inducible p53DINP1|stress-induced protein|tumor protein p53-inducible nuclear protein 1 |
Position | 8q22 |
Gene Type | protein-coding |
Mutation summary: | Mutations in domains Mutational pattern in 17 major cancer types LoF vs missense mutations All the copy number variations from COSMIC database V72 All the mutations from COSMIC database V72 |
Mutation summary in protein domains | Top |
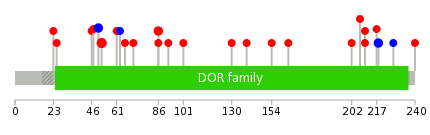 |
The non-synonymous (top) and synonymous (bottom) mutational percentage for various cancers | Top |
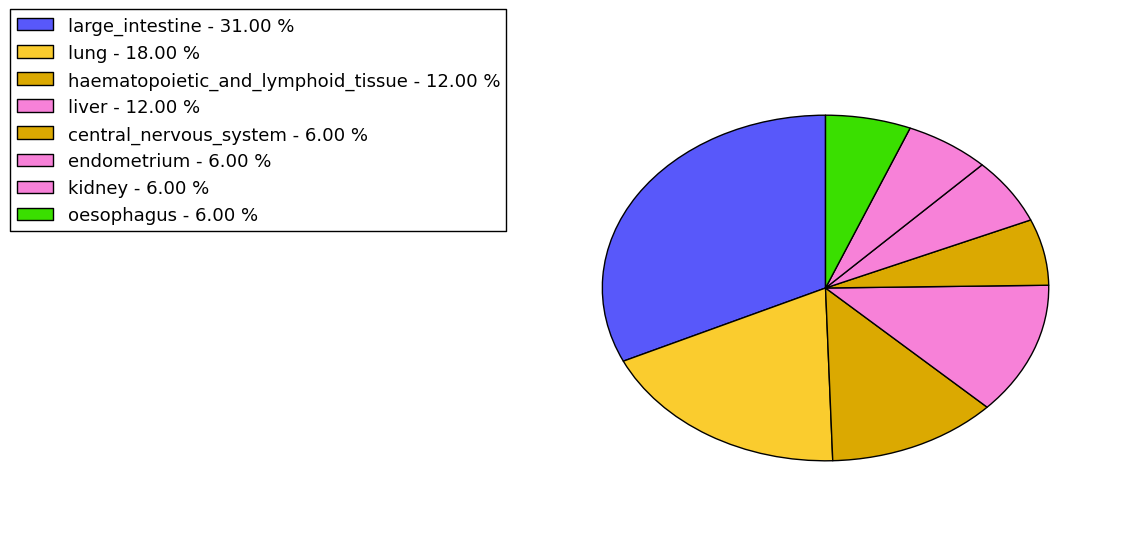 |
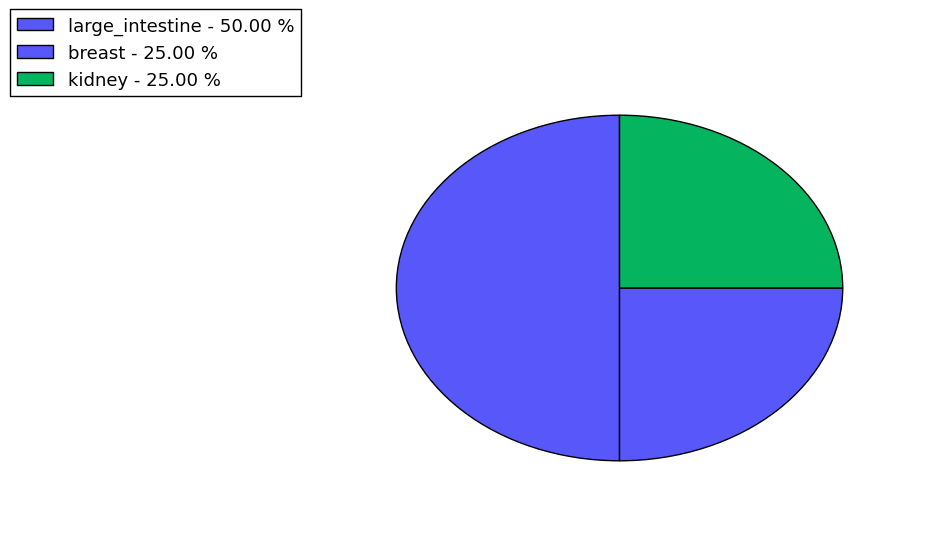 |
Loss of Function mutations compare to missense mutations | Top |
 |
| The ratio of the loss-of-function mutations over missense mutations is 0.12. |
The copy number variations for various cancers | Top |
 |
The copy number variations in 17 major cancer types: 1; Breast, 2; Central nervous system, 3; Cervix, 4; Endometrium, 5; Haematopoietic and lymphoid_tissue, 6; Kidney, 7; Large intestine, 8; Liver, 9; Lung, 10; Not specific, 11; Ovary, 12; Pancreas, 13; Prostate, 14; Skin, 15; Stomach, 16; Thyroid, 17; Urinary tract |
COSMIC somatic mutation [Top] | |||
|
Mutation (CDS; AA; Chr) |
Site |
Histology |
Mutation Type |
|---|---|---|---|
c.150A>G; p.E50E; 8:94940183-94940183 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.150A>G; p.E50E; 8:94940183-94940183 |
breast | carcinoma | Substitution - coding silent |
c.681C>A; p.G227G; 8:94930521-94930521 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - coding silent |
c.681C>A; p.G227G; 8:94930521-94930521 |
large_intestine | carcinoma; adenocarcinoma | Substitution - coding silent |
c.416C>A; p.P139H; 8:94939917-94939917 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - Missense |
c.196T>C; p.F66L; 8:94940137-94940137 |
central_nervous_system; brain | glioma; astrocytoma_Grade_IV | Substitution - Missense |
c.181G>A; p.E61K; 8:94940152-94940152 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.276G>T; p.M92I; 8:94940057-94940057 |
oesophagus | carcinoma; adenocarcinoma | Substitution - Missense |
c.189T>C; p.P63P; 8:94940144-94940144 |
skin | malignant_melanoma | Substitution - coding silent |
c.621T>G; p.N207K; 8:94930581-94930581 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; chronic_lymphocytic_leukaemia-small_lymphocytic_lymphoma | Substitution - Missense |
c.257A>G; p.Q86R; 8:94940076-94940076 |
liver | carcinoma | Substitution - Missense |
c.257A>G; p.Q86R; 8:94940076-94940076 |
liver | carcinoma; hepatocellular_carcinoma | Substitution - Missense |
c.150_152delAGA; p.E52delE; 8:94940181-94940183 |
skin | malignant_melanoma | Deletion - In frame |
c.150_152delAGA; p.E52delE; 8:94940181-94940183 |
urinary_tract; bladder | carcinoma; transitional_cell_carcinoma | Deletion - In frame |
c.150_152delAGA; p.E52delE; 8:94940181-94940183 |
endometrium | carcinoma; endometrioid_carcinoma | Deletion - In frame |
c.150_152delAGA; p.E52delE; 8:94940181-94940183 |
stomach | carcinoma; intestinal_adenocarcinoma | Deletion - In frame |
c.718T>A; p.Y240N; 8:94930484-94930484 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.606G>T; p.Q202H; 8:94930596-94930596 |
large_intestine | carcinoma; adenocarcinoma | Substitution - Missense |
c.137A>C; p.E46A; 8:94940196-94940196 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.605A>G; p.Q202R; 8:94930597-94930597 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.490G>A; p.E164K; 8:94930712-94930712 |
skin; trunk | malignant_melanoma; nodular | Substitution - Missense |
c.211G>A; p.A71T; 8:94940122-94940122 |
stomach | carcinoma; adenocarcinoma | Substitution - Missense |
c.73G>C; p.E25Q; 8:94940869-94940869 |
urinary_tract; bladder | carcinoma | Substitution - Missense |
c.139G>A; p.E47K; 8:94940194-94940194 |
large_intestine; caecum | carcinoma; adenocarcinoma | Substitution - Missense |
c.628C>T; p.R210C; 8:94930574-94930574 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.654C>T; p.C218C; 8:94930548-94930548 |
kidney | carcinoma; clear_cell_renal_cell_carcinoma | Substitution - coding silent |
c.654C>T; p.C218C; 8:94930548-94930548 |
prostate | carcinoma | Substitution - coding silent |
c.461C>A; p.P154Q; 8:94939872-94939872 |
skin | malignant_melanoma | Substitution - Missense |
c.649G>A; p.D217N; 8:94930553-94930553 |
large_intestine; colon | carcinoma; adenocarcinoma | Substitution - Missense |
c.301C>T; p.P101S; 8:94940032-94940032 |
thyroid | carcinoma | Substitution - Missense |
c.389C>G; p.S130C; 8:94939944-94939944 |
haematopoietic_and_lymphoid_tissue | haematopoietic_neoplasm; acute_myeloid_leukaemia | Substitution - Missense |
c.256C>T; p.Q86*; 8:94940077-94940077 |
haematopoietic_and_lymphoid_tissue | lymphoid_neoplasm; acute_lymphoblastic_leukaemia | Substitution - Nonsense |
c.589G>T; p.E197*; 8:94930613-94930613 |
large_intestine; rectum | carcinoma; adenocarcinoma | Substitution - Nonsense |
c.69C>A; p.F23L; 8:94940873-94940873 |
endometrium | carcinoma; endometrioid_carcinoma | Substitution - Missense |
c.256C>G; p.Q86E; 8:94940077-94940077 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |
c.629G>A; p.R210H; 8:94930573-94930573 |
lung | carcinoma; adenocarcinoma | Substitution - Missense |