| General information | Literature | Expression | Regulation | Variant | Interaction |
|
Basic Information |
|
|---|---|
Gene ID | 142 |
Name | PARP1 |
Synonymous | ADPRT|ADPRT 1|ADPRT1|ARTD1|PARP|PARP-1|PPOL|pADPRT-1;poly (ADP-ribose) polymerase 1;PARP1;poly (ADP-ribose) polymerase 1 |
Definition | ADP-ribosyltransferase (NAD+; poly (ADP-ribose) polymerase)|ADP-ribosyltransferase NAD(+)|ADP-ribosyltransferase diphtheria toxin-like 1|NAD(+) ADP-ribosyltransferase 1|poly (ADP-ribose) polymerase family, member 1|poly [ADP-ribose] polymerase 1|poly(ADP- |
Position | 1q41-q42 |
Gene Type | protein-coding |
Gene Regulation: | Upstream microRNAs Transcription Factors Post Transcriptional Modification From dbPTM Methylation Profile From DiseaseMeth database |
Experimentally verified miRNA from miRTarBase [Top] | |||
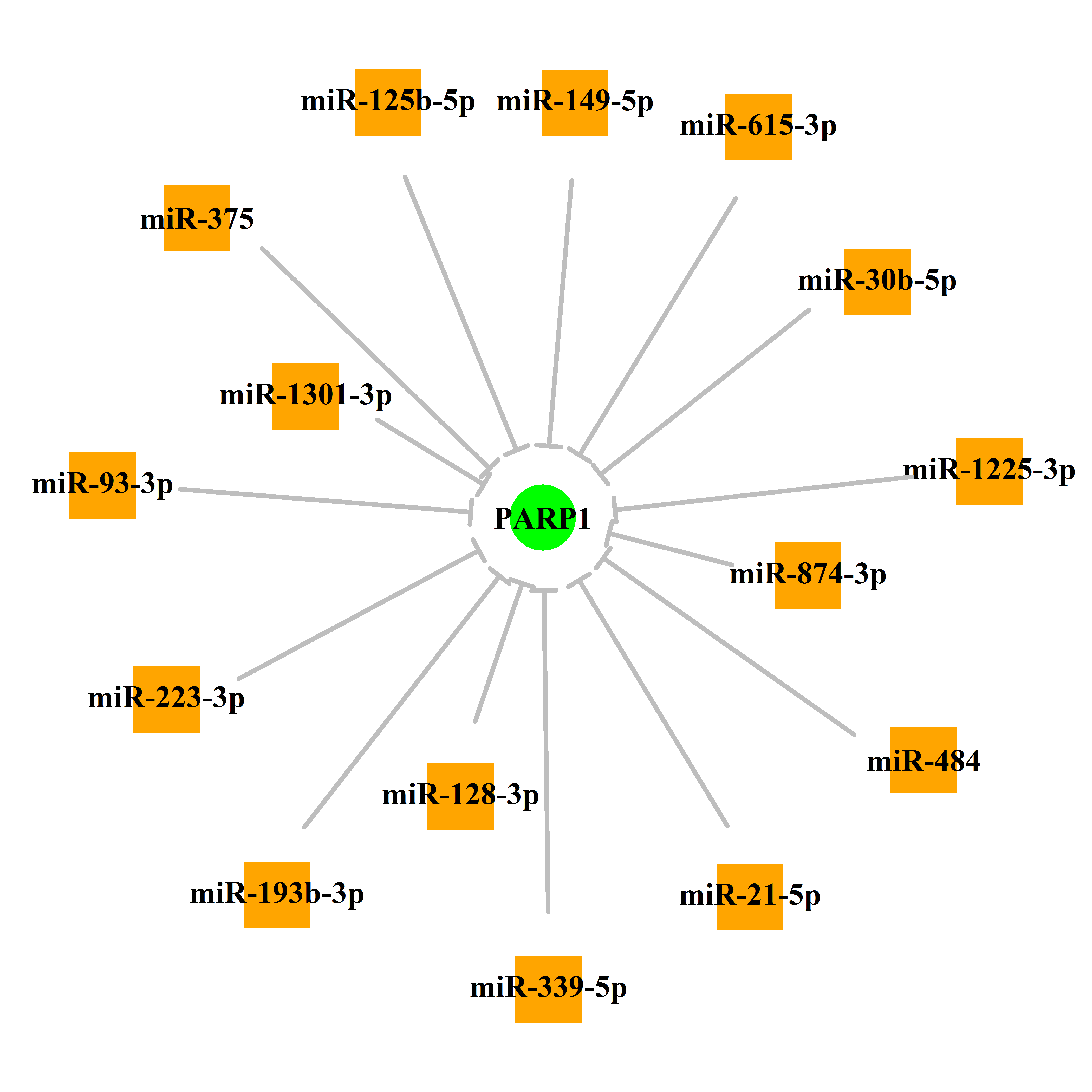 |
Transcription Factors [Top] |
|
Regulation From TransFac database |
PARP(h) --Caspase-3(h)--> PARP-p24(h) + PARP-p89(h) (processing) | PARP(h) --Caspase-7(h)--> PARP-p24(h) + PARP-p89(h) (processing) | PARP(h) + Ku autoantigen(h) <==> PARP(h):Ku autoantigen(h) (binding) | PARP(h) + DNA-PK(h) <==> PARP(h):DNA-PK(h) (binding) | PARP(h) --> PARP-p89(h) + protein remnants (cleavage) | PARP(h) --Caspase-3(h)--> protein remnants (cleavage) | PARP(h) --Caspase-8(h)--> protein remnants (cleavage) | PARP(h) + YY1(h) <==> PARP(h):YY1(h) (binding) | PARP(h) + NAD --PARP(h)--> PARP(h){drib} + nicotinamide (ADP-ribosylation) | PARP(h) --> protein remnants (cleavage) | PCNA(h) + PARP(h) <==> PCNA(h):PARP(h) (binding) | p21Cip1(h) + PARP(h) <==> p21Cip1(h):PARP(h) (binding) | PARP(h) --> PARP-p85(h) + protein remnants (cleavage) | TERF2IP(h) + TRF2(h) + Mre11(h) + Ku70(h) + Ku80(h) + PARP(h) + Rad50(h) <==> TERF2IP(h):TRF2(h):Mre11(h):Ku70(h):Ku80(h):PARP(h):Rad50(h) (binding) | PARP(h) + TERF2IP(h) <==> PARP(h):TERF2IP(h) (binding) | PARP(h) + AR-isoform1(h) <==> PARP(h):AR-isoform1(h) (binding) | PARP(h) + AR(h) <==> PARP(h):AR(h) (binding) | PARP(h) --Caspase-3(h)--> PARP-p85(h) + protein remnants (cleavage) | PARP(h) --Caspase-3(m.s.)--> PARP-p85(h) + protein remnants (cleavage) | PARP(h) --> PARP-p25(h) + protein remnants (cleavage) | p300(h) + PARP(h) <==> p300(h):PARP(h) (binding) | p300(h) + PARP(h) <==> p300(h):PARP(h) (binding) | PARP(h) + acetyl-CoA --p300(h)--> PARP(h){aceK} + CoA (acetylation) | PARP(h) + acetyl-CoA --CBP(h)--> PARP(h){aceK} + CoA (acetylation) | PARP(h) + acetyl-CoA --> PARP(h){aceK} + CoA (acetylation) | Cdk8(h) + MED7(m.s.) + TRAP80(m.s.) + DRIP100(m.s.) + DRIP130(m.s.) + TRAP170(m.s.) + DRIP205(m.s.) + TFIIF-alpha(m.s.) + p300(h) + PARP(h) <==> Cdk8(h):MED7(m.s.):TRAP80(m.s.):DRIP100(m.s.):DRIP130(m.s.):TRAP170(m.s.):DRIP205(m.s.):TFIIF-alpha(m.s.):p300(h):PARP(h) (binding) | RelA-p65(h) + PARP(h) <==> RelA-p65(h):PARP(h) (binding) | NF-kappaB1(h) + PARP(h) <==> NF-kappaB1(h):PARP(h) (binding) | HDAC1(m.s.) + PARP(h) <==> HDAC1(m.s.):PARP(h) (binding) | PARP(h) + hdac2(m.s.) <==> PARP(h):hdac2(m.s.) (binding) | PARP(h) + HDAC3(m.s.) <==> PARP(h):HDAC3(m.s.) (binding) | PARP(h) --> SNCA(h) (DNA binding). | TCF-4(h) + PARP(h) <==> TCF-4(h):PARP(h) (binding) | Ku70(h) + PARP(h) <==> Ku70(h):PARP(h) (binding) | Ku80(h) + PARP(h) <==> Ku80(h):PARP(h) (binding) | NURR1(m.s.) + PARP(h) <==> NURR1(m.s.):PARP(h) (binding) | PARP(h) + NOR1(m.s.) <==> PARP(h):NOR1(m.s.) (binding) | PARP(h) + NOR1(h) <==> PARP(h):NOR1(h) (binding) | PARP(h) + TRAP170(h) <==> PARP(h):TRAP170(h) (binding) | PARP(h) --> CXCL1(h) (DNA binding). | PARP(h) --> PARP-p89(h) + protein remnants (processing) | PARP(h) --> PARP-p29(h) + protein remnants (cleavage) | PARP(h) + BRRN1(m.s.) <==> PARP(h):BRRN1(m.s.) (binding) | PARP(h) + BRRN1(h) <==> PARP(h):BRRN1(h) (binding) | PARP(h) + xrcc1(h) <==> PARP(h):xrcc1(h) (binding) | FEN1(h) + PARP(h) <==> FEN1(h):PARP(h) (binding) | PARP(h) + NAD --> PARP(h){drib} + nicotinamide (ADP-ribosylation; automodification) | PARP(h) --> VIM(h) (DNA binding). | PARP1(h) --> PARP(h) (expression). | HCV-core-1B(HCV1B) --> PARP(h) (increase of cleavage) | NADE-isoform1(m) --> PARP(h) (increase of cleavage) | YY1(h) + NAD --PARP(h)--> YY1(h){drib} + nicotinamide (ADP-ribosylation) | PARP(h) + NAD --PARP(h)--> PARP(h){drib} + nicotinamide (ADP-ribosylation) | TBP(h):TFIIB(h) + NAD --PARP(h)--> TBP(h){drib}:TFIIB(h) + nicotinamide (ADP-ribosylation) | Max(h) + PARP(h) <==> Max(h):PARP(h) (binding) | PARP(h) + TEF-1(r) <==> PARP(h):TEF-1(r) (binding) | PARP(h) + TTF-1(r) <==> PARP(h):TTF-1(r) (binding) | PARP(h) --> SP-B(m) (transactivation) | PARP(h) + NAD --> PARP(h){drib} + nicotinamide (ADP-ribosylation; automodification) | PARP(h) + 5 acetyl-CoA --p300(h)--> PARP(h){aceK498}{aceK505}{aceK508}{aceK521}{aceK524} + 5 CoA (acetylation) | PARP(h) + 5 acetyl-CoA --CBP(h)--> PARP(h){aceK498}{aceK505}{aceK508}{aceK521}{aceK524} + 5 CoA (acetylation) | PARP(h) + Cdk8(m.s.) <==> PARP(h):Cdk8(m.s.) (binding) | PARP(h) + TRAP170(m.s.) <==> PARP(h):TRAP170(m.s.) (binding) | PARP(h) + HMGIY(m.s.) <==> PARP(h):HMGIY(m.s.) (binding) | PARP(h) + TFIIF-alpha(m.s.) <==> PARP(h):TFIIF-alpha(m.s.) (binding) | p300(h) + PARP(h) <==> p300(h):PARP(h) (binding) | 2 PARP(h) <==> (PARP(h))2 (oligomerization) | PARP(h) + RelA-p65(m.s.) <==> PARP(h):RelA-p65(m.s.) (binding) | PARP(h) + NF-kappaB1(m.s.) <==> PARP(h):NF-kappaB1(m.s.) (binding) | PARP(h) + cyclinC(h) <==> PARP(h):cyclinC(h) (binding) | PARP(h) + HDAC3(m.s.) <==> PARP(h):HDAC3(m.s.) (binding) | PARP(h) + hdac2(m.s.) <==> PARP(h):hdac2(m.s.) (binding) | PARP(h) + HDAC1(m.s.) <==> PARP(h):HDAC1(m.s.) (binding) | PARP(h) + Cdk8(h) <==> PARP(h):Cdk8(h) (binding) | PARP(h) --> SNCA(h) (DNA binding). | TCF-4(h) + PARP(h) <==> TCF-4(h):PARP(h) (binding) | PARP(h) --> VIM(h) (transactivation) | PARP1(h) --> PARP(h) (expression). |
|---|
Post Transcriptional Modification [Top] | |||
|
Location (AA) |
PTM type |
Literature |
Database |
407 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 413 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 435 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 437 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 444 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 445 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 448 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 456 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 471 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 484 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 488 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 491 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 513 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 514 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 | 520 | PolyADP-ribosyl glutamic acid(Potential). | Swiss-Prot 53.0 |
|---|---|---|---|
Methylation Profile [Top] | ||
|
Chromosome |
Location |
Source |
|---|---|---|
chr1 |
Promoter: 224661914 - 224663914 | Agilent_014791 |
chr1 |
Promoter: 224661914 - 224663914 | GoldenGate_Methylation_Cancer_Panel_I |
chr1 |
Promoter: 224661914 - 224663914 | GSE27584 |
chr1 |
Promoter: 224661914 - 224663914 | Agilent_014791_44K |