rs10997361
Homo sapiens
C>T
CTNNA3 : Intron Variant
SNV (Single Nucleotide Variation)
T=0261 (7837/29930,GnomAD)
T=0210 (6133/29118,TOPMED)
T=0381 (1906/5008,1000G)
T=0256 (986/3854,ALSPAC)
T=0250 (927/3708,TWINSUK)
T=0210 (6133/29118,TOPMED)
T=0381 (1906/5008,1000G)
T=0256 (986/3854,ALSPAC)
T=0250 (927/3708,TWINSUK)
chr10:66722696 (GRCh38.p7) (10q21.3)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 10 | NC_000010.11:g.66722696C>T |
| GRCh37.p13 chr 10 | NC_000010.10:g.68482454C>T |
| CTNNA3 RefSeqGene | NG_034072.1:g.978496G>A |
Gene: CTNNA3, catenin alpha 3(minus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| CTNNA3 transcript variant 2 | NM_001127384.2:c. | N/A | Intron Variant |
| CTNNA3 transcript variant 1 | NM_013266.3:c. | N/A | Intron Variant |
| CTNNA3 transcript variant 3 | NM_001291133.1:c. | N/A | Genic Downstream Transcript Variant |
| CTNNA3 transcript variant X1 | XM_017016151.1:c. | N/A | Intron Variant |
| CTNNA3 transcript variant X2 | XM_017016152.1:c. | N/A | Intron Variant |
| CTNNA3 transcript variant X3 | XM_017016153.1:c. | N/A | Intron Variant |
| CTNNA3 transcript variant X4 | XM_017016154.1:c. | N/A | Intron Variant |
| CTNNA3 transcript variant X5 | XM_017016155.1:c. | N/A | Intron Variant |
| CTNNA3 transcript variant X6 | XM_017016156.1:c. | N/A | Intron Variant |
| CTNNA3 transcript variant X7 | XM_017016157.1:c. | N/A | Intron Variant |
| CTNNA3 transcript variant X8 | XM_017016158.1:c. | N/A | Genic Downstream Transcript Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | C=0.934 | T=0.066 |
| 1000Genomes | American | Sub | 694 | C=0.500 | T=0.500 |
| 1000Genomes | East Asian | Sub | 1008 | C=0.208 | T=0.792 |
| 1000Genomes | Europe | Sub | 1006 | C=0.702 | T=0.298 |
| 1000Genomes | Global | Study-wide | 5008 | C=0.619 | T=0.381 |
| 1000Genomes | South Asian | Sub | 978 | C=0.620 | T=0.380 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | C=0.744 | T=0.256 |
| The Genome Aggregation Database | African | Sub | 8716 | C=0.917 | T=0.083 |
| The Genome Aggregation Database | American | Sub | 836 | C=0.500 | T=0.500 |
| The Genome Aggregation Database | East Asian | Sub | 1612 | C=0.179 | T=0.821 |
| The Genome Aggregation Database | Europe | Sub | 18464 | C=0.714 | T=0.285 |
| The Genome Aggregation Database | Global | Study-wide | 29930 | C=0.738 | T=0.261 |
| The Genome Aggregation Database | Other | Sub | 302 | C=0.660 | T=0.340 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | C=0.789 | T=0.210 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | C=0.750 | T=0.250 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 20201924 | Genome-wide association study of alcohol dependence implicates a region on chromosome 11. | Edenberg HJ | Alcohol Clin Exp Res |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs10997361 | 0.000294 | alcohol dependence | 20201924 |
eQTL of rs10997361 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs10997361 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
Genomic View
Chromatin Interaction
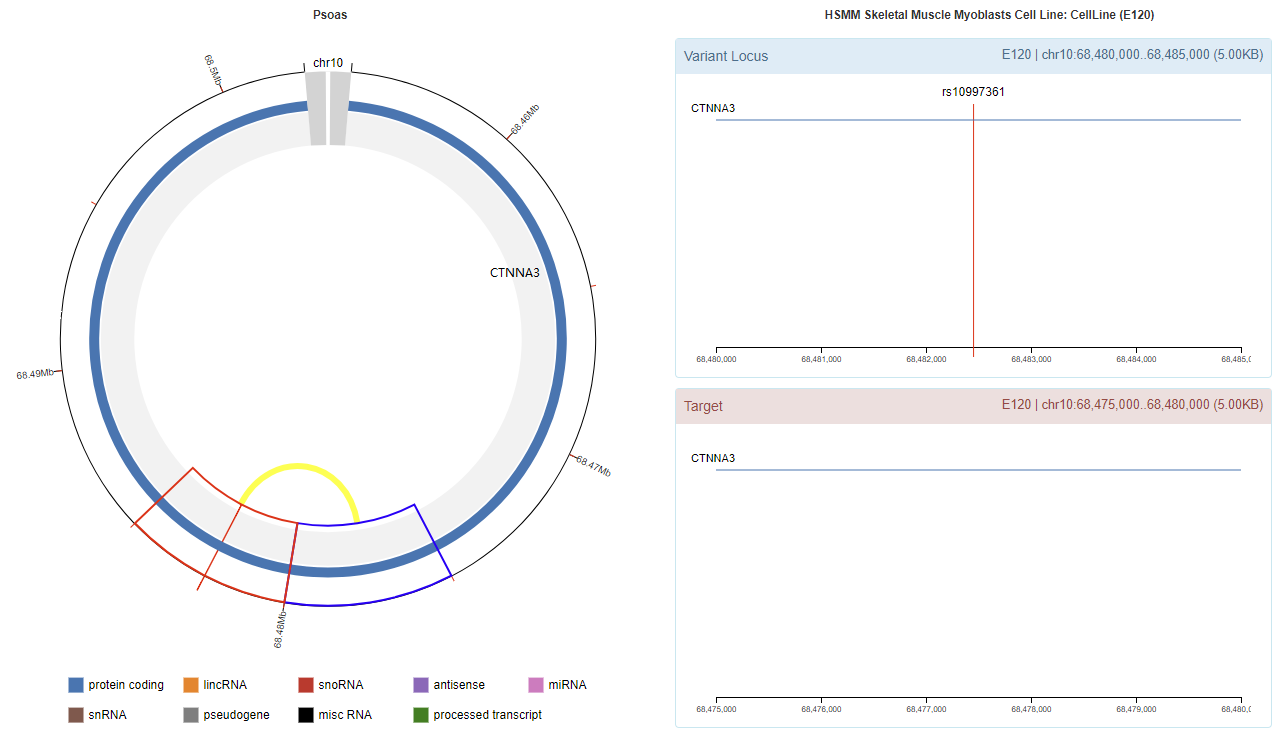
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr10 | 68483530 | 68483599 | E067 | 1076 |
| chr10 | 68483530 | 68483599 | E069 | 1076 |
| chr10 | 68483695 | 68483990 | E069 | 1241 |
| chr10 | 68484110 | 68484288 | E070 | 1656 |
| chr10 | 68483530 | 68483599 | E071 | 1076 |
| chr10 | 68483695 | 68483990 | E071 | 1241 |
| chr10 | 68484110 | 68484288 | E071 | 1656 |
| chr10 | 68483530 | 68483599 | E072 | 1076 |
| chr10 | 68483695 | 68483990 | E072 | 1241 |
| chr10 | 68484110 | 68484288 | E072 | 1656 |
| chr10 | 68506369 | 68506750 | E081 | 23915 |
| chr10 | 68483530 | 68483599 | E082 | 1076 |
| chr10 | 68483695 | 68483990 | E082 | 1241 |
| chr10 | 68520439 | 68521142 | E082 | 37985 |