rs11200700
Homo sapiens
A>G
None
SNV (Single Nucleotide Variation)
G=0161 (4847/29950,GnomAD)
G=0164 (4784/29118,TOPMED)
G=0138 (689/5008,1000G)
G=0155 (599/3854,ALSPAC)
G=0163 (605/3708,TWINSUK)
G=0164 (4784/29118,TOPMED)
G=0138 (689/5008,1000G)
G=0155 (599/3854,ALSPAC)
G=0163 (605/3708,TWINSUK)
chr10:83854079 (GRCh38.p7) (10q23.1)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 10 | NC_000010.11:g.83854079A>G |
| GRCh37.p13 chr 10 | NC_000010.10:g.85613835A>G |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | A=0.778 | G=0.222 |
| 1000Genomes | American | Sub | 694 | A=0.910 | G=0.090 |
| 1000Genomes | East Asian | Sub | 1008 | A=0.904 | G=0.096 |
| 1000Genomes | Europe | Sub | 1006 | A=0.832 | G=0.168 |
| 1000Genomes | Global | Study-wide | 5008 | A=0.862 | G=0.138 |
| 1000Genomes | South Asian | Sub | 978 | A=0.930 | G=0.070 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | A=0.845 | G=0.155 |
| The Genome Aggregation Database | African | Sub | 8710 | A=0.785 | G=0.215 |
| The Genome Aggregation Database | American | Sub | 838 | A=0.890 | G=0.110 |
| The Genome Aggregation Database | East Asian | Sub | 1614 | A=0.889 | G=0.111 |
| The Genome Aggregation Database | Europe | Sub | 18486 | A=0.856 | G=0.143 |
| The Genome Aggregation Database | Global | Study-wide | 29950 | A=0.838 | G=0.161 |
| The Genome Aggregation Database | Other | Sub | 302 | A=0.850 | G=0.150 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | A=0.835 | G=0.164 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | A=0.837 | G=0.163 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 20201924 | Genome-wide association study of alcohol dependence implicates a region on chromosome 11. | Edenberg HJ | Alcohol Clin Exp Res |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs11200700 | 0.0001 | alcoholism | pha002891 |
| rs11200700 | 0.0001 | alcohol dependence | 20201924 |
eQTL of rs11200700 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs11200700 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
Genomic View
Chromatin Interaction
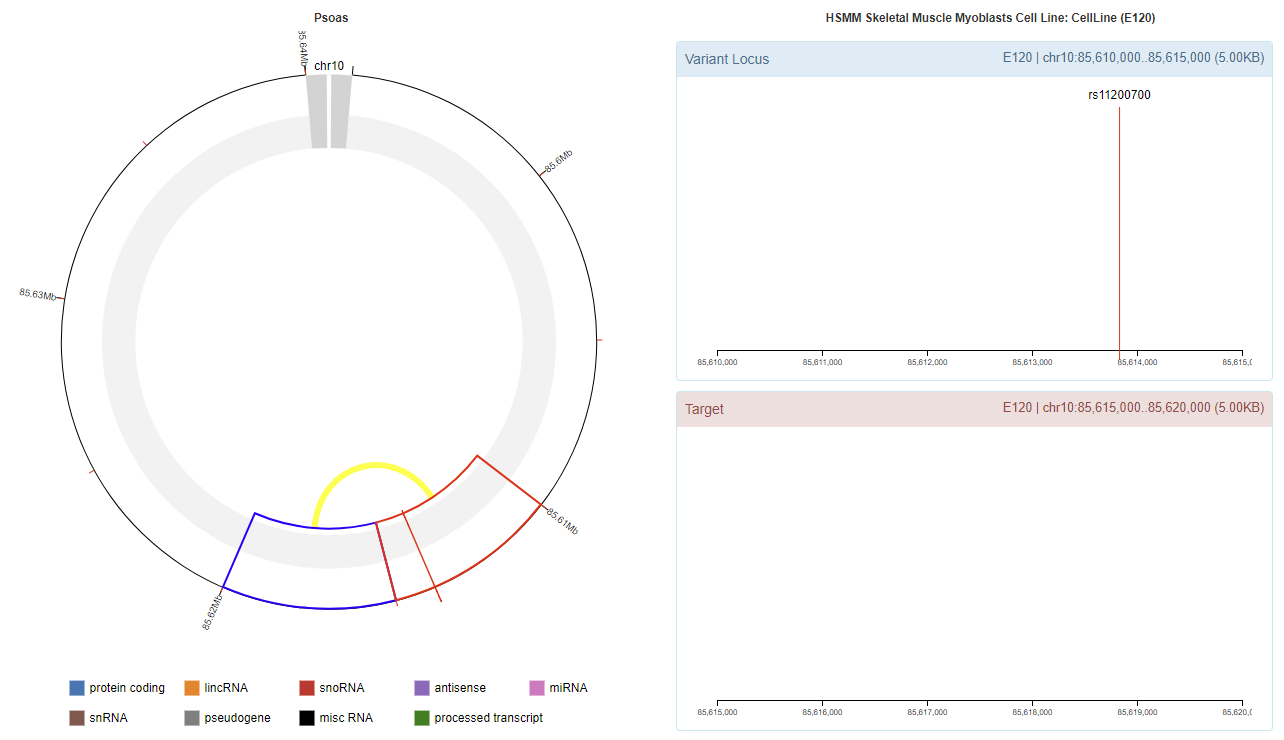
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr10 | 85646574 | 85646626 | E068 | 32739 |
| chr10 | 85646741 | 85646939 | E068 | 32906 |
| chr10 | 85641366 | 85641807 | E069 | 27531 |
| chr10 | 85635600 | 85635703 | E070 | 21765 |
| chr10 | 85635890 | 85636108 | E070 | 22055 |
| chr10 | 85640986 | 85641316 | E070 | 27151 |
| chr10 | 85641366 | 85641807 | E070 | 27531 |
| chr10 | 85646089 | 85646204 | E070 | 32254 |
| chr10 | 85646271 | 85646475 | E070 | 32436 |
| chr10 | 85646574 | 85646626 | E070 | 32739 |
| chr10 | 85646741 | 85646939 | E070 | 32906 |
| chr10 | 85640986 | 85641316 | E071 | 27151 |
| chr10 | 85641366 | 85641807 | E071 | 27531 |
| chr10 | 85630797 | 85630847 | E072 | 16962 |
| chr10 | 85630905 | 85630965 | E072 | 17070 |
| chr10 | 85630971 | 85631058 | E072 | 17136 |
| chr10 | 85583678 | 85583728 | E081 | -30107 |
| chr10 | 85583770 | 85583828 | E081 | -30007 |
| chr10 | 85583860 | 85583910 | E081 | -29925 |
| chr10 | 85584013 | 85584122 | E081 | -29713 |
| chr10 | 85584172 | 85584252 | E081 | -29583 |
| chr10 | 85584302 | 85584428 | E081 | -29407 |
| chr10 | 85646089 | 85646204 | E082 | 32254 |
| chr10 | 85646271 | 85646475 | E082 | 32436 |