rs11717973
Homo sapiens
C>T
ITGA9 : Intron Variant
SNV (Single Nucleotide Variation)
T=0469 (14039/29902,GnomAD)
T=0448 (13070/29118,TOPMED)
C==0478 (2396/5008,1000G)
C==0495 (1906/3854,ALSPAC)
C==0492 (1825/3708,TWINSUK)
T=0448 (13070/29118,TOPMED)
C==0478 (2396/5008,1000G)
C==0495 (1906/3854,ALSPAC)
C==0492 (1825/3708,TWINSUK)
chr3:37720523 (GRCh38.p7) (3p22.2)
ND
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 3 | NC_000003.12:g.37720523C>T |
| GRCh37.p13 chr 3 | NC_000003.11:g.37762014C>T |
| ITGA9 RefSeqGene | NG_016166.1:g.273202C>T |
Gene: ITGA9, integrin subunit alpha 9(plus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| ITGA9 transcript | NM_002207.2:c. | N/A | Intron Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | C=0.617 | T=0.383 |
| 1000Genomes | American | Sub | 694 | C=0.490 | T=0.510 |
| 1000Genomes | East Asian | Sub | 1008 | C=0.436 | T=0.564 |
| 1000Genomes | Europe | Sub | 1006 | C=0.516 | T=0.484 |
| 1000Genomes | Global | Study-wide | 5008 | C=0.478 | T=0.522 |
| 1000Genomes | South Asian | Sub | 978 | C=0.290 | T=0.710 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | C=0.495 | T=0.505 |
| The Genome Aggregation Database | African | Sub | 8704 | C=0.585 | T=0.415 |
| The Genome Aggregation Database | American | Sub | 834 | C=0.450 | T=0.550 |
| The Genome Aggregation Database | East Asian | Sub | 1608 | C=0.467 | T=0.533 |
| The Genome Aggregation Database | Europe | Sub | 18454 | C=0.513 | T=0.486 |
| The Genome Aggregation Database | Global | Study-wide | 29902 | C=0.530 | T=0.469 |
| The Genome Aggregation Database | Other | Sub | 302 | C=0.560 | T=0.440 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | C=0.551 | T=0.448 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | C=0.492 | T=0.508 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 19268276 | Genome-wide association study of smoking initiation and current smoking. | Vink JM | Am J Hum Genet |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs11717973 | 0.000476 | nicotine smoking | 19268276 |
eQTL of rs11717973 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs11717973 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
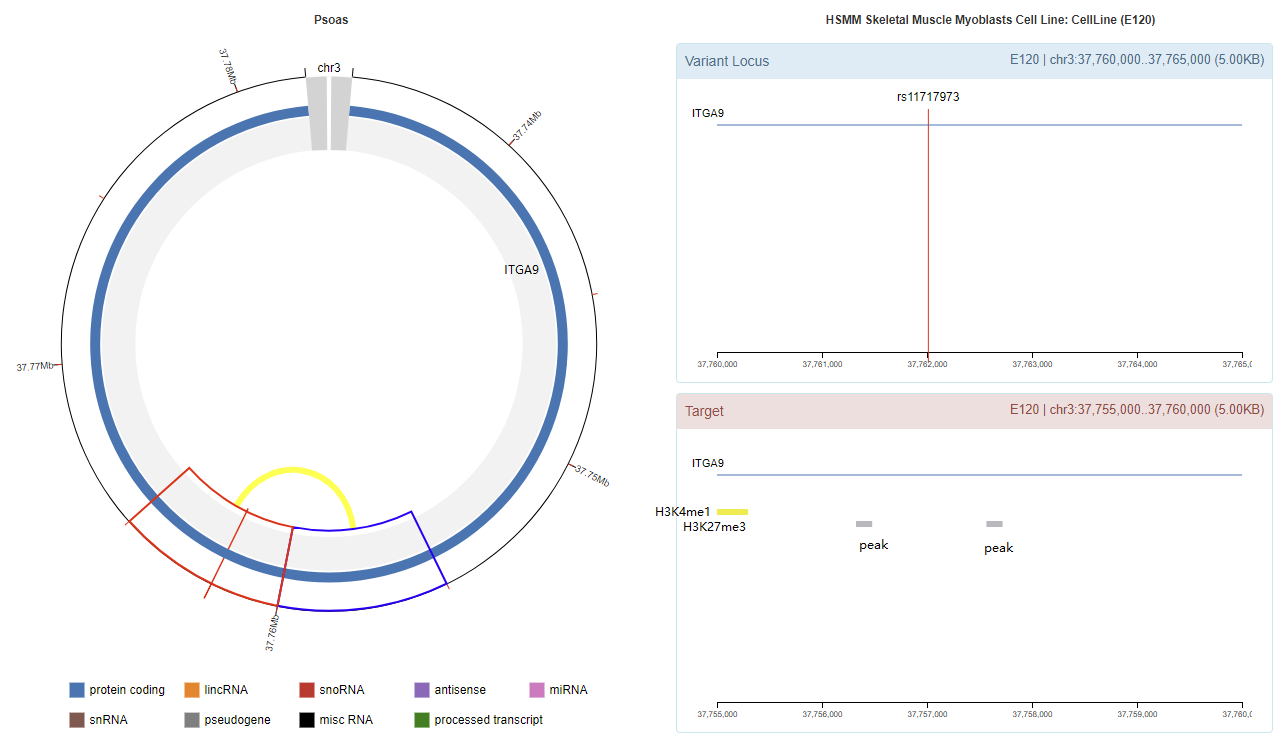
Genomic View
Chromatin Interaction
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr3 | 37735124 | 37736312 | E067 | -25702 |
| chr3 | 37736416 | 37736535 | E067 | -25479 |
| chr3 | 37783231 | 37783432 | E067 | 21217 |
| chr3 | 37731361 | 37731509 | E068 | -30505 |
| chr3 | 37754498 | 37754615 | E068 | -7399 |
| chr3 | 37754633 | 37755280 | E068 | -6734 |
| chr3 | 37804709 | 37805445 | E068 | 42695 |
| chr3 | 37735124 | 37736312 | E069 | -25702 |
| chr3 | 37736416 | 37736535 | E069 | -25479 |
| chr3 | 37754006 | 37754289 | E069 | -7725 |
| chr3 | 37754498 | 37754615 | E069 | -7399 |
| chr3 | 37754633 | 37755280 | E069 | -6734 |
| chr3 | 37755282 | 37755369 | E069 | -6645 |
| chr3 | 37804709 | 37805445 | E069 | 42695 |
| chr3 | 37805494 | 37805619 | E069 | 43480 |
| chr3 | 37745095 | 37745448 | E070 | -16566 |
| chr3 | 37745528 | 37745690 | E070 | -16324 |
| chr3 | 37721699 | 37722589 | E071 | -39425 |
| chr3 | 37735124 | 37736312 | E071 | -25702 |
| chr3 | 37736416 | 37736535 | E071 | -25479 |
| chr3 | 37754633 | 37755280 | E071 | -6734 |
| chr3 | 37755282 | 37755369 | E071 | -6645 |
| chr3 | 37755474 | 37755527 | E071 | -6487 |
| chr3 | 37783231 | 37783432 | E071 | 21217 |
| chr3 | 37804709 | 37805445 | E071 | 42695 |
| chr3 | 37805494 | 37805619 | E071 | 43480 |
| chr3 | 37735124 | 37736312 | E072 | -25702 |
| chr3 | 37736416 | 37736535 | E072 | -25479 |
| chr3 | 37736747 | 37736942 | E072 | -25072 |
| chr3 | 37755474 | 37755527 | E072 | -6487 |
| chr3 | 37804709 | 37805445 | E072 | 42695 |
| chr3 | 37805494 | 37805619 | E072 | 43480 |
| chr3 | 37754498 | 37754615 | E073 | -7399 |
| chr3 | 37754633 | 37755280 | E073 | -6734 |
| chr3 | 37755282 | 37755369 | E073 | -6645 |
| chr3 | 37804709 | 37805445 | E073 | 42695 |
| chr3 | 37805494 | 37805619 | E073 | 43480 |
| chr3 | 37735124 | 37736312 | E074 | -25702 |
| chr3 | 37736416 | 37736535 | E074 | -25479 |
| chr3 | 37754498 | 37754615 | E074 | -7399 |
| chr3 | 37754633 | 37755280 | E074 | -6734 |
| chr3 | 37755282 | 37755369 | E074 | -6645 |
| chr3 | 37755474 | 37755527 | E074 | -6487 |
| chr3 | 37804709 | 37805445 | E074 | 42695 |
| chr3 | 37805494 | 37805619 | E074 | 43480 |
| chr3 | 37730751 | 37730857 | E081 | -31157 |
| chr3 | 37735124 | 37736312 | E081 | -25702 |
| chr3 | 37754633 | 37755280 | E081 | -6734 |