rs180172
Homo sapiens
A>G
LOC107983956 : Intron Variant
SNV (Single Nucleotide Variation)
G=0096 (2889/29956,GnomAD)
G=0097 (2835/29118,TOPMED)
G=0121 (607/5008,1000G)
G=0122 (472/3854,ALSPAC)
G=0116 (430/3708,TWINSUK)
G=0097 (2835/29118,TOPMED)
G=0121 (607/5008,1000G)
G=0122 (472/3854,ALSPAC)
G=0116 (430/3708,TWINSUK)
chr17:70041173 (GRCh38.p7) (17q24.3)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 17 | NC_000017.11:g.70041173A>G |
| GRCh37.p13 chr 17 | NC_000017.10:g.68037314A>G |
Gene: LOC107983956, uncharacterized LOC107983956(minus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| LOC105371881 transcript | XR_934952.2:n. | N/A | Intron Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | A=0.889 | G=0.111 |
| 1000Genomes | American | Sub | 694 | A=0.870 | G=0.130 |
| 1000Genomes | East Asian | Sub | 1008 | A=0.994 | G=0.006 |
| 1000Genomes | Europe | Sub | 1006 | A=0.902 | G=0.098 |
| 1000Genomes | Global | Study-wide | 5008 | A=0.879 | G=0.121 |
| 1000Genomes | South Asian | Sub | 978 | A=0.730 | G=0.270 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | A=0.878 | G=0.122 |
| The Genome Aggregation Database | African | Sub | 8708 | A=0.898 | G=0.102 |
| The Genome Aggregation Database | American | Sub | 838 | A=0.860 | G=0.140 |
| The Genome Aggregation Database | East Asian | Sub | 1620 | A=0.995 | G=0.005 |
| The Genome Aggregation Database | Europe | Sub | 18488 | A=0.899 | G=0.100 |
| The Genome Aggregation Database | Global | Study-wide | 29956 | A=0.903 | G=0.096 |
| The Genome Aggregation Database | Other | Sub | 302 | A=0.930 | G=0.070 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | A=0.902 | G=0.097 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | A=0.884 | G=0.116 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 21314694 | Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. | Kendler KS | Alcohol Clin Exp Res |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs180172 | 0.000648 | alcohol dependence | 21314694 |
eQTL of rs180172 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs180172 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
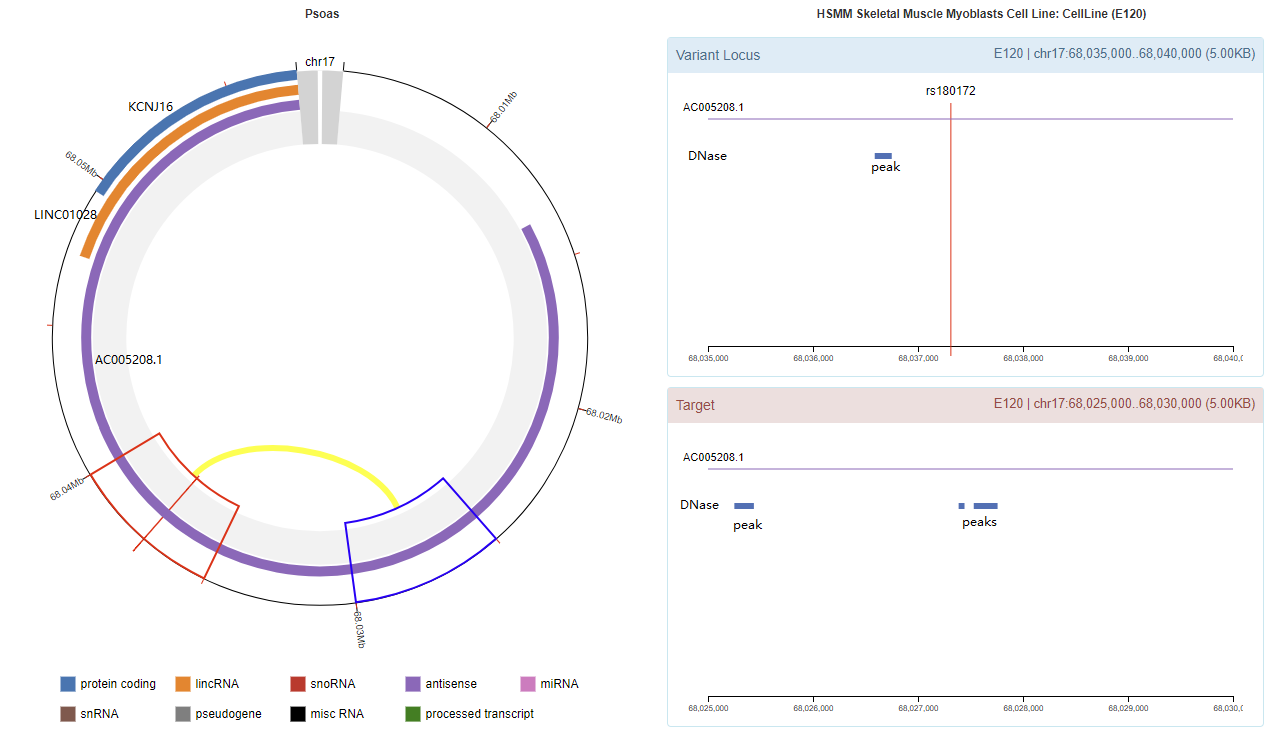
Genomic View
Chromatin Interaction
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr17 | 68063079 | 68063172 | E068 | 25765 |
| chr17 | 68063347 | 68063758 | E068 | 26033 |
| chr17 | 68063905 | 68063955 | E068 | 26591 |
| chr17 | 68063079 | 68063172 | E069 | 25765 |
| chr17 | 68063347 | 68063758 | E069 | 26033 |
| chr17 | 68063905 | 68063955 | E069 | 26591 |
| chr17 | 68069171 | 68069334 | E069 | 31857 |
| chr17 | 68069409 | 68069578 | E069 | 32095 |
| chr17 | 68069619 | 68069741 | E069 | 32305 |
| chr17 | 68069762 | 68069819 | E069 | 32448 |
| chr17 | 68069923 | 68069970 | E069 | 32609 |
| chr17 | 68069923 | 68069970 | E070 | 32609 |
| chr17 | 68070859 | 68071558 | E070 | 33545 |
| chr17 | 68071598 | 68071663 | E070 | 34284 |
| chr17 | 68063347 | 68063758 | E071 | 26033 |
| chr17 | 68069171 | 68069334 | E071 | 31857 |
| chr17 | 68069409 | 68069578 | E071 | 32095 |
| chr17 | 68063347 | 68063758 | E072 | 26033 |
| chr17 | 68069171 | 68069334 | E072 | 31857 |
| chr17 | 68069409 | 68069578 | E072 | 32095 |
| chr17 | 68063079 | 68063172 | E074 | 25765 |
| chr17 | 68063347 | 68063758 | E074 | 26033 |
| chr17 | 68063905 | 68063955 | E074 | 26591 |
| chr17 | 68069171 | 68069334 | E074 | 31857 |
| chr17 | 68069409 | 68069578 | E074 | 32095 |
| chr17 | 68069619 | 68069741 | E074 | 32305 |
| chr17 | 68069762 | 68069819 | E074 | 32448 |
| chr17 | 68069923 | 68069970 | E074 | 32609 |
| chr17 | 68070060 | 68070234 | E074 | 32746 |
| chr17 | 68056325 | 68056425 | E081 | 19011 |
| chr17 | 68056495 | 68056554 | E081 | 19181 |
| chr17 | 68056657 | 68056828 | E081 | 19343 |