rs2674052
Homo sapiens
T>A / T>G
None
SNV (Single Nucleotide Variation)
G=0295 (8843/29960,GnomAD)
G=0363 (1819/5008,1000G)
G=0321 (1236/3854,ALSPAC)
G=0324 (1202/3708,TWINSUK)
G=0363 (1819/5008,1000G)
G=0321 (1236/3854,ALSPAC)
G=0324 (1202/3708,TWINSUK)
chr2:59543218 (GRCh38.p7) (2p16.1)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 2 | NC_000002.12:g.59543218T>A |
| GRCh38.p7 chr 2 | NC_000002.12:g.59543218T>G |
| GRCh37.p13 chr 2 | NC_000002.11:g.59770353T>A |
| GRCh37.p13 chr 2 | NC_000002.11:g.59770353T>G |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | T=0.737 | G=0.263 |
| 1000Genomes | American | Sub | 694 | T=0.590 | G=0.410 |
| 1000Genomes | East Asian | Sub | 1008 | T=0.472 | G=0.528 |
| 1000Genomes | Europe | Sub | 1006 | T=0.677 | G=0.323 |
| 1000Genomes | Global | Study-wide | 5008 | T=0.637 | G=0.363 |
| 1000Genomes | South Asian | Sub | 978 | T=0.660 | G=0.340 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | T=0.679 | G=0.321 |
| The Genome Aggregation Database | African | Sub | 8724 | T=0.737 | G=0.263 |
| The Genome Aggregation Database | American | Sub | 838 | T=0.570 | G=0.43, |
| The Genome Aggregation Database | East Asian | Sub | 1614 | T=0.468 | G=0.532 |
| The Genome Aggregation Database | Europe | Sub | 18482 | T=0.717 | G=0.282 |
| The Genome Aggregation Database | Global | Study-wide | 29960 | T=0.704 | G=0.295 |
| The Genome Aggregation Database | Other | Sub | 302 | T=0.660 | G=0.34, |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | T=0.676 | G=0.324 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 23743675 | A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. | Kapoor M | Hum Genet |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs2674052 | 6.82E-05 | alcohol consumption | 23743675 |
eQTL of rs2674052 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs2674052 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value |
|---|---|---|---|---|
| cg03558837 | chr2:239029375 | ESPNL | 0.0646226347905888 | 2.4442e-14 |
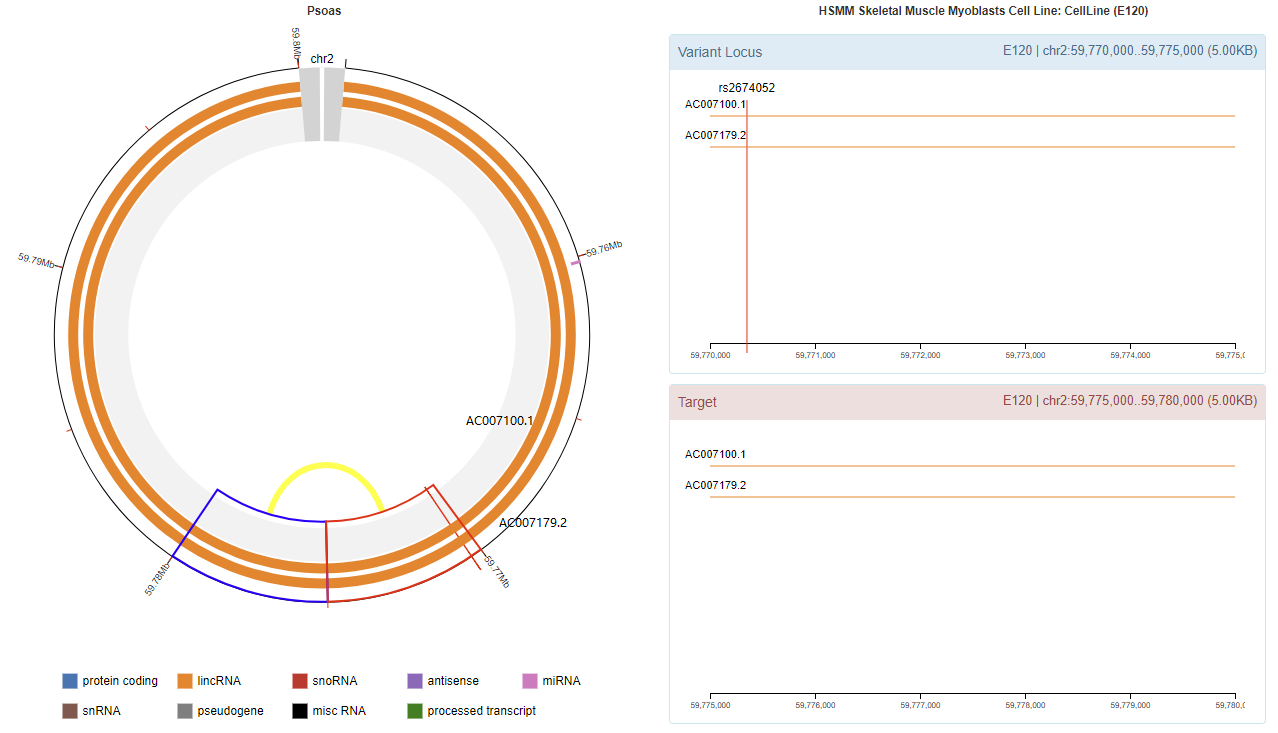
Genomic View
Chromatin Interaction
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr2 | 238950342 | 238950447 | E067 | -42598 |
| chr2 | 238951505 | 238951913 | E067 | -41132 |
| chr2 | 238970839 | 238970899 | E067 | -22146 |
| chr2 | 238990205 | 238990255 | E067 | -2790 |
| chr2 | 238990452 | 238990751 | E067 | -2294 |
| chr2 | 238970839 | 238970899 | E068 | -22146 |
| chr2 | 239017313 | 239017876 | E068 | 24268 |
| chr2 | 238951505 | 238951913 | E069 | -41132 |
| chr2 | 238970839 | 238970899 | E069 | -22146 |
| chr2 | 238989790 | 238989866 | E069 | -3179 |
| chr2 | 238989941 | 238990032 | E069 | -3013 |
| chr2 | 238990205 | 238990255 | E069 | -2790 |
| chr2 | 238970839 | 238970899 | E070 | -22146 |
| chr2 | 238950342 | 238950447 | E071 | -42598 |
| chr2 | 238951505 | 238951913 | E071 | -41132 |
| chr2 | 238951961 | 238952020 | E071 | -41025 |
| chr2 | 238970839 | 238970899 | E071 | -22146 |
| chr2 | 238989247 | 238989354 | E071 | -3691 |
| chr2 | 238989790 | 238989866 | E071 | -3179 |
| chr2 | 238989941 | 238990032 | E071 | -3013 |
| chr2 | 238990205 | 238990255 | E071 | -2790 |
| chr2 | 238990452 | 238990751 | E071 | -2294 |
| chr2 | 239007116 | 239007529 | E071 | 14071 |
| chr2 | 239017176 | 239017226 | E071 | 24131 |
| chr2 | 239017313 | 239017876 | E071 | 24268 |
| chr2 | 238950342 | 238950447 | E072 | -42598 |
| chr2 | 238989790 | 238989866 | E072 | -3179 |
| chr2 | 238989941 | 238990032 | E072 | -3013 |
| chr2 | 238990205 | 238990255 | E072 | -2790 |
| chr2 | 238990452 | 238990751 | E072 | -2294 |
| chr2 | 239014417 | 239014467 | E072 | 21372 |
| chr2 | 239014951 | 239015001 | E072 | 21906 |
| chr2 | 238970839 | 238970899 | E073 | -22146 |
| chr2 | 239014951 | 239015001 | E073 | 21906 |
| chr2 | 238950342 | 238950447 | E074 | -42598 |
| chr2 | 238951505 | 238951913 | E074 | -41132 |
| chr2 | 238989790 | 238989866 | E074 | -3179 |
| chr2 | 238989941 | 238990032 | E074 | -3013 |
| chr2 | 238990452 | 238990751 | E074 | -2294 |
| chr2 | 239017313 | 239017876 | E074 | 24268 |
| chr2 | 238994008 | 238994058 | E081 | 963 |
| chr2 | 238994372 | 238994803 | E081 | 1327 |
| chr2 | 238993565 | 238993671 | E082 | 520 |
| chr2 | 238994008 | 238994058 | E082 | 963 |
Promoter Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance(-/+:Up/Downstream) |
|---|---|---|---|---|
| chr2 | 238968700 | 238970607 | E067 | -22438 |
| chr2 | 238968700 | 238970607 | E068 | -22438 |
| chr2 | 238968700 | 238970607 | E069 | -22438 |
| chr2 | 238968700 | 238970607 | E070 | -22438 |
| chr2 | 238968700 | 238970607 | E071 | -22438 |
| chr2 | 238968700 | 238970607 | E072 | -22438 |
| chr2 | 238968700 | 238970607 | E073 | -22438 |
| chr2 | 238968700 | 238970607 | E074 | -22438 |
| chr2 | 238968700 | 238970607 | E081 | -22438 |
| chr2 | 238968700 | 238970607 | E082 | -22438 |