rs2837437
Homo sapiens
T>C
DSCAM : Intron Variant
SNV (Single Nucleotide Variation)
C=0106 (3176/29964,GnomAD)
C=0114 (3331/29118,TOPMED)
C=0106 (530/5008,1000G)
C=0102 (392/3854,ALSPAC)
C=0095 (351/3708,TWINSUK)
C=0114 (3331/29118,TOPMED)
C=0106 (530/5008,1000G)
C=0102 (392/3854,ALSPAC)
C=0095 (351/3708,TWINSUK)
chr21:40126099 (GRCh38.p7) (21q22.2)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 21 | NC_000021.9:g.40126099T>C |
| GRCh37.p13 chr 21 | NC_000021.8:g.41498026T>C |
Gene: DSCAM, Down syndrome cell adhesion molecule(minus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| DSCAM transcript variant 2 | NM_001271534.1:c. | N/A | Intron Variant |
| DSCAM transcript variant 1 | NM_001389.3:c. | N/A | Intron Variant |
| DSCAM transcript variant 3 | NR_073202.1:n. | N/A | Intron Variant |
| DSCAM transcript variant X1 | XM_017028281.1:c. | N/A | Intron Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | T=0.858 | C=0.142 |
| 1000Genomes | American | Sub | 694 | T=0.930 | C=0.070 |
| 1000Genomes | East Asian | Sub | 1008 | T=0.861 | C=0.139 |
| 1000Genomes | Europe | Sub | 1006 | T=0.904 | C=0.096 |
| 1000Genomes | Global | Study-wide | 5008 | T=0.894 | C=0.106 |
| 1000Genomes | South Asian | Sub | 978 | T=0.940 | C=0.060 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | T=0.898 | C=0.102 |
| The Genome Aggregation Database | African | Sub | 8726 | T=0.848 | C=0.152 |
| The Genome Aggregation Database | American | Sub | 836 | T=0.940 | C=0.060 |
| The Genome Aggregation Database | East Asian | Sub | 1620 | T=0.877 | C=0.123 |
| The Genome Aggregation Database | Europe | Sub | 18480 | T=0.914 | C=0.085 |
| The Genome Aggregation Database | Global | Study-wide | 29964 | T=0.894 | C=0.106 |
| The Genome Aggregation Database | Other | Sub | 302 | T=0.910 | C=0.090 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | T=0.885 | C=0.114 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | T=0.905 | C=0.095 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 20201924 | Genome-wide association study of alcohol dependence implicates a region on chromosome 11. | Edenberg HJ | Alcohol Clin Exp Res |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs2837437 | 0.00081 | alcohol dependence | 20201924 |
eQTL of rs2837437 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs2837437 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
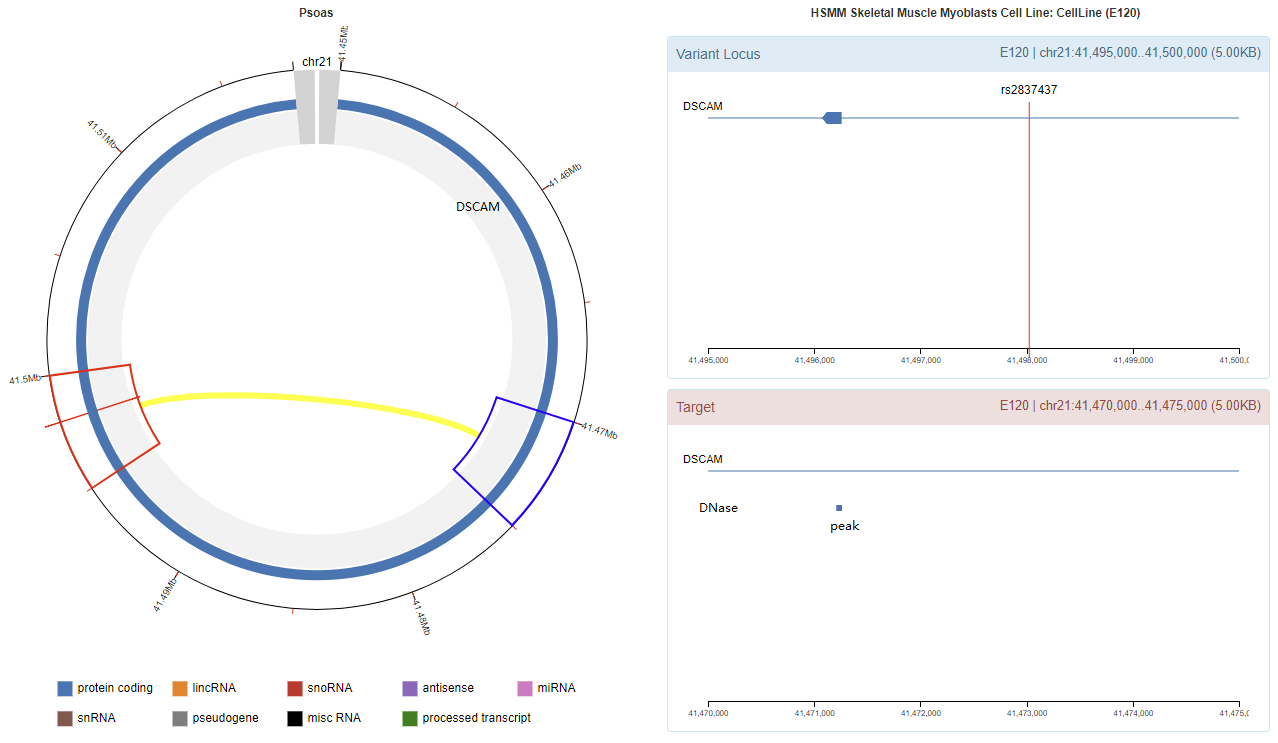
Genomic View
Chromatin Interaction
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr21 | 41492849 | 41493177 | E067 | -4849 |
| chr21 | 41538341 | 41538454 | E067 | 40315 |
| chr21 | 41538485 | 41538651 | E067 | 40459 |
| chr21 | 41506211 | 41506353 | E068 | 8185 |
| chr21 | 41506211 | 41506353 | E069 | 8185 |
| chr21 | 41526615 | 41526665 | E070 | 28589 |
| chr21 | 41526767 | 41526863 | E070 | 28741 |
| chr21 | 41526978 | 41527190 | E070 | 28952 |
| chr21 | 41532941 | 41533072 | E070 | 34915 |
| chr21 | 41506211 | 41506353 | E072 | 8185 |
| chr21 | 41452431 | 41452634 | E081 | -45392 |
| chr21 | 41452710 | 41452760 | E081 | -45266 |
| chr21 | 41452898 | 41453192 | E081 | -44834 |
| chr21 | 41516283 | 41516434 | E081 | 18257 |
| chr21 | 41516680 | 41516730 | E081 | 18654 |
| chr21 | 41526615 | 41526665 | E082 | 28589 |
| chr21 | 41526767 | 41526863 | E082 | 28741 |
| chr21 | 41526978 | 41527190 | E082 | 28952 |