rs368028
Homo sapiens
C>T
None
SNV (Single Nucleotide Variation)
C==0180 (5406/29944,GnomAD)
C==0141 (4123/29118,TOPMED)
C==0272 (1362/5008,1000G)
C==0158 (608/3854,ALSPAC)
C==0152 (562/3708,TWINSUK)
C==0141 (4123/29118,TOPMED)
C==0272 (1362/5008,1000G)
C==0158 (608/3854,ALSPAC)
C==0152 (562/3708,TWINSUK)
chr6:169411199 (GRCh38.p7) (6q27)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 6 | NC_000006.12:g.169411199C>T |
| GRCh37.p13 chr 6 | NC_000006.11:g.169811294C>T |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | C=0.089 | T=0.911 |
| 1000Genomes | American | Sub | 694 | C=0.310 | T=0.690 |
| 1000Genomes | East Asian | Sub | 1008 | C=0.592 | T=0.408 |
| 1000Genomes | Europe | Sub | 1006 | C=0.160 | T=0.840 |
| 1000Genomes | Global | Study-wide | 5008 | C=0.272 | T=0.728 |
| 1000Genomes | South Asian | Sub | 978 | C=0.280 | T=0.720 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | C=0.158 | T=0.842 |
| The Genome Aggregation Database | African | Sub | 8718 | C=0.104 | T=0.896 |
| The Genome Aggregation Database | American | Sub | 838 | C=0.340 | T=0.660 |
| The Genome Aggregation Database | East Asian | Sub | 1614 | C=0.615 | T=0.385 |
| The Genome Aggregation Database | Europe | Sub | 18474 | C=0.172 | T=0.827 |
| The Genome Aggregation Database | Global | Study-wide | 29944 | C=0.180 | T=0.819 |
| The Genome Aggregation Database | Other | Sub | 300 | C=0.160 | T=0.840 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | C=0.141 | T=0.858 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | C=0.152 | T=0.848 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 21314694 | Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. | Kendler KS | Alcohol Clin Exp Res |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs368028 | 0.000911 | alcohol dependence | 21314694 |
eQTL of rs368028 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue |
|---|---|---|---|---|---|---|
| Chr6:169811294 | RP1-137D17.1 | ENSG00000226194.1 | C>T | 1.3823e-4 | 22814 | Cerebellar_Hemisphere |
| Chr6:169811294 | RP1-137D17.1 | ENSG00000226194.1 | C>T | 9.3356e-4 | 22814 | Caudate_basal_ganglia |
meQTL of rs368028 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
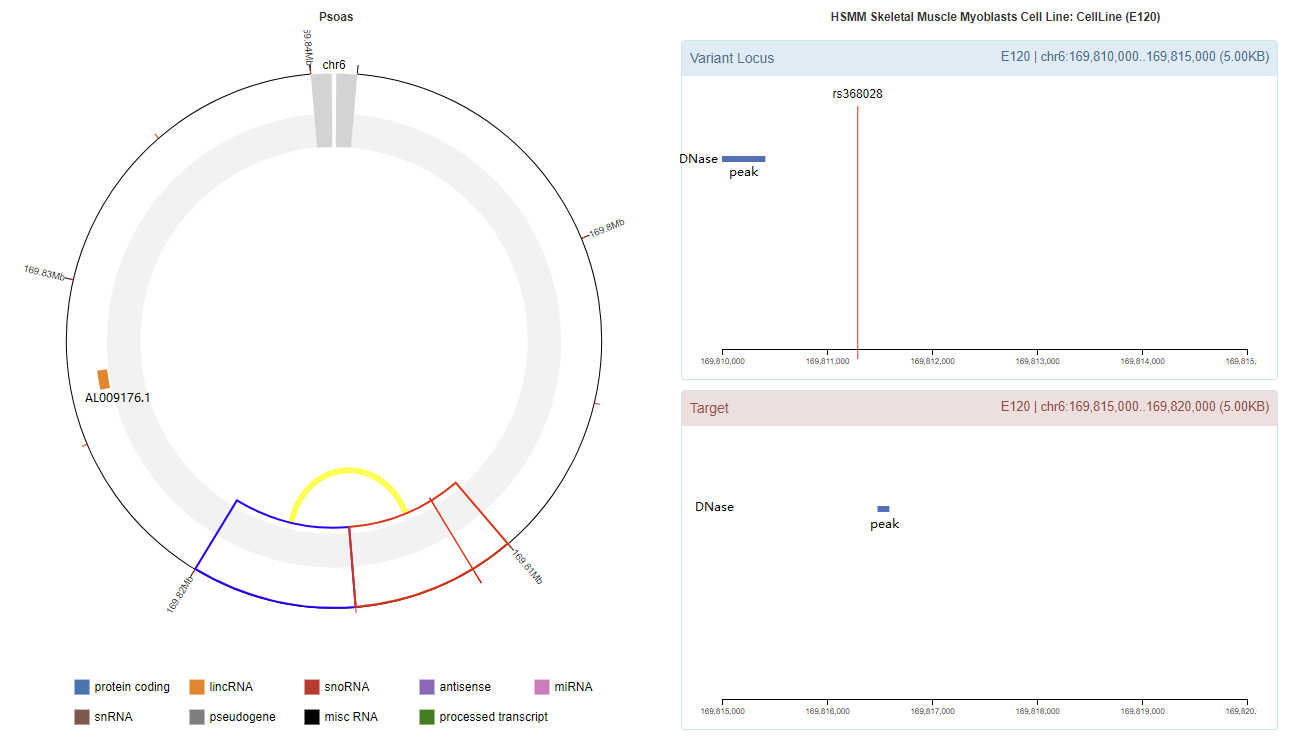
Genomic View
Chromatin Interaction
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr6 | 169762720 | 169763070 | E068 | -48224 |
| chr6 | 169765748 | 169766004 | E069 | -45290 |
| chr6 | 169766117 | 169766167 | E069 | -45127 |
| chr6 | 169827198 | 169828587 | E070 | 15904 |
| chr6 | 169828658 | 169829133 | E070 | 17364 |
| chr6 | 169850221 | 169850528 | E070 | 38927 |
| chr6 | 169850536 | 169850627 | E070 | 39242 |
| chr6 | 169850767 | 169850891 | E070 | 39473 |
| chr6 | 169850221 | 169850528 | E071 | 38927 |
| chr6 | 169850536 | 169850627 | E071 | 39242 |
| chr6 | 169850767 | 169850891 | E071 | 39473 |
| chr6 | 169765748 | 169766004 | E072 | -45290 |
| chr6 | 169766117 | 169766167 | E072 | -45127 |
| chr6 | 169766241 | 169766338 | E072 | -44956 |
| chr6 | 169766388 | 169766448 | E072 | -44846 |
| chr6 | 169777940 | 169778203 | E082 | -33091 |
| chr6 | 169778760 | 169778855 | E082 | -32439 |
| chr6 | 169828658 | 169829133 | E082 | 17364 |
Promoter Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance(-/+:Up/Downstream) |
|---|---|---|---|---|
| chr6 | 169852002 | 169852614 | E067 | 40708 |
| chr6 | 169852002 | 169852614 | E071 | 40708 |