rs3924047
Homo sapiens
T>C
SHANK2 : Intron Variant
SNV (Single Nucleotide Variation)
C=0493 (14761/29894,GnomAD)
C=0482 (14059/29118,TOPMED)
T==0474 (6772/14288,ExAC)
T==0481 (2407/5008,1000G)
T==0477 (2179/4566,GO-ESP)
C=0495 (1907/3854,ALSPAC)
C=0481 (1784/3708,TWINSUK)
C=0482 (14059/29118,TOPMED)
T==0474 (6772/14288,ExAC)
T==0481 (2407/5008,1000G)
T==0477 (2179/4566,GO-ESP)
C=0495 (1907/3854,ALSPAC)
C=0481 (1784/3708,TWINSUK)
chr11:71118812 (GRCh38.p7) (11q13.4)
AD
GWASdb2
Genomic Coordinates
| Sequence Name | Change(s) |
|---|---|
| GRCh38.p7 chr 11 | NC_000011.10:g.71118812T>C |
| GRCh37.p13 chr 11 fix patch HG865_PATCH | NW_004070871.1:g.613025T>C |
| GRCh37.p13 chr 11 | NC_000011.9:g.70829858T>C |
Gene: SHANK2, SH3 and multiple ankyrin repeat domains 2(minus strand)
| Molecule type | Change | Amino acid[Codon] | SO Term |
|---|---|---|---|
| SHANK2 transcript variant 1 | NM_012309.4:c. | N/A | Intron Variant |
| SHANK2 transcript variant 2 | NM_133266.4:c. | N/A | Genic Upstream Transcript Variant |
| SHANK2 transcript variant 3 | NR_110766.1:n. | N/A | Genic Upstream Transcript Variant |
| SHANK2 transcript variant X1 | XM_017017387.1:c. | N/A | Intron Variant |
| SHANK2 transcript variant X2 | XM_017017388.1:c. | N/A | Intron Variant |
| SHANK2 transcript variant X3 | XM_017017389.1:c. | N/A | Intron Variant |
| SHANK2 transcript variant X4 | XM_005277932.3:c. | N/A | Genic Upstream Transcript Variant |
| SHANK2 transcript variant X5 | XM_017017390.1:c. | N/A | Genic Upstream Transcript Variant |
Population Frequency
| Study | Population | Group | Sample # | Ref Allele | Alt Allele |
|---|---|---|---|---|---|
| 1000Genomes | African | Sub | 1322 | T=0.482 | C=0.518 |
| 1000Genomes | American | Sub | 694 | T=0.440 | C=0.560 |
| 1000Genomes | East Asian | Sub | 1008 | T=0.384 | C=0.616 |
| 1000Genomes | Europe | Sub | 1006 | T=0.500 | C=0.500 |
| 1000Genomes | Global | Study-wide | 5008 | T=0.481 | C=0.519 |
| 1000Genomes | South Asian | Sub | 978 | T=0.580 | C=0.420 |
| The Avon Longitudinal Study of Parents and Children | PARENT AND CHILD COHORT | Study-wide | 3854 | T=0.505 | C=0.495 |
| The Exome Aggregation Consortium | American | Sub | 2428 | T=0.500 | C=0.500 |
| The Exome Aggregation Consortium | Asian | Sub | 3754 | T=0.446 | C=0.554 |
| The Exome Aggregation Consortium | Europe | Sub | 7964 | T=0.479 | C=0.521 |
| The Exome Aggregation Consortium | Global | Study-wide | 14288 | T=0.474 | C=0.526 |
| The Exome Aggregation Consortium | Other | Sub | 142 | T=0.450 | C=0.550 |
| The Genome Aggregation Database | African | Sub | 8706 | T=0.521 | C=0.479 |
| The Genome Aggregation Database | American | Sub | 834 | T=0.450 | C=0.550 |
| The Genome Aggregation Database | East Asian | Sub | 1612 | T=0.389 | C=0.611 |
| The Genome Aggregation Database | Europe | Sub | 18440 | T=0.510 | C=0.489 |
| The Genome Aggregation Database | Global | Study-wide | 29894 | T=0.506 | C=0.493 |
| The Genome Aggregation Database | Other | Sub | 302 | T=0.600 | C=0.400 |
| Trans-Omics for Precision Medicine | Global | Study-wide | 29118 | T=0.517 | C=0.482 |
| UK 10K study - Twins | TWIN COHORT | Study-wide | 3708 | T=0.519 | C=0.481 |
| PMID | Title | Author | Journal |
|---|---|---|---|
| 16400611 | Genomewide linkage scan of 409 European-ancestry and African American families with schizophrenia: suggestive evidence of linkage at 8p23.3-p21.2 and 11p13.1-q14.1 in the combined sample. | Suarez BK | Am J Hum Genet |
| 23691058 | Dosage transmission disequilibrium test (dTDT) for linkage and association detection. | Zhang Z | PLoS One |
P-Value
| SNP ID | p-value | Traits | Study |
|---|---|---|---|
| rs3924047 | 8.77E-06 | alcohol dependence | 23691058 |
eQTL of rs3924047 in Brain tissues (GTEx Analysis Release V7)
| Position (v37) | eGene | GeneID | Variant | p-value | TSS | Tissue | |
|---|---|---|---|---|---|---|---|
| There is no eQTL annotation for this SNP | |||||||
meQTL of rs3924047 in Fetal Brain
| Probe ID | Position | Gene | beta | p-value | |||
|---|---|---|---|---|---|---|---|
| There is no meQTL annotation for this SNP | |||||||
Genomic View
Chromatin Interaction
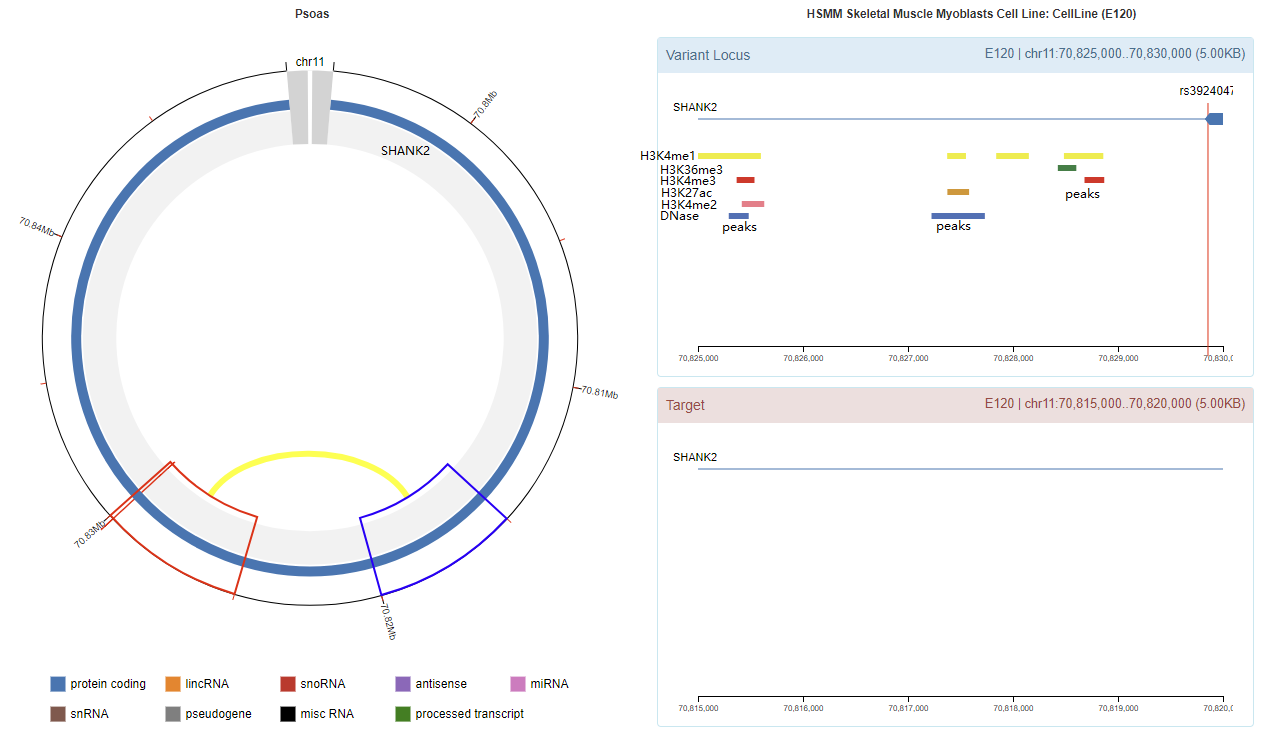
Enhancer Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance ( -/+ : Up/Downstream ) |
|---|---|---|---|---|
| chr11 | 578622 | 578923 | E067 | -34102 |
| chr11 | 579295 | 579830 | E067 | -33195 |
| chr11 | 579896 | 579951 | E067 | -33074 |
| chr11 | 580027 | 580591 | E067 | -32434 |
| chr11 | 581922 | 581972 | E067 | -31053 |
| chr11 | 632579 | 632671 | E067 | 19554 |
| chr11 | 632853 | 633265 | E067 | 19828 |
| chr11 | 571203 | 572078 | E068 | -40947 |
| chr11 | 572115 | 572210 | E068 | -40815 |
| chr11 | 578622 | 578923 | E068 | -34102 |
| chr11 | 579058 | 579102 | E068 | -33923 |
| chr11 | 579186 | 579249 | E068 | -33776 |
| chr11 | 579295 | 579830 | E068 | -33195 |
| chr11 | 579896 | 579951 | E068 | -33074 |
| chr11 | 580027 | 580591 | E068 | -32434 |
| chr11 | 581922 | 581972 | E068 | -31053 |
| chr11 | 614155 | 614458 | E068 | 1130 |
| chr11 | 658284 | 658350 | E068 | 45259 |
| chr11 | 658432 | 658924 | E068 | 45407 |
| chr11 | 578622 | 578923 | E069 | -34102 |
| chr11 | 579058 | 579102 | E069 | -33923 |
| chr11 | 579186 | 579249 | E069 | -33776 |
| chr11 | 579295 | 579830 | E069 | -33195 |
| chr11 | 579896 | 579951 | E069 | -33074 |
| chr11 | 580027 | 580591 | E069 | -32434 |
| chr11 | 614155 | 614458 | E069 | 1130 |
| chr11 | 632579 | 632671 | E069 | 19554 |
| chr11 | 632853 | 633265 | E069 | 19828 |
| chr11 | 658284 | 658350 | E069 | 45259 |
| chr11 | 569960 | 570100 | E070 | -42925 |
| chr11 | 570113 | 570215 | E070 | -42810 |
| chr11 | 578622 | 578923 | E070 | -34102 |
| chr11 | 579058 | 579102 | E070 | -33923 |
| chr11 | 579186 | 579249 | E070 | -33776 |
| chr11 | 579295 | 579830 | E070 | -33195 |
| chr11 | 579896 | 579951 | E070 | -33074 |
| chr11 | 580027 | 580591 | E070 | -32434 |
| chr11 | 580655 | 580709 | E070 | -32316 |
| chr11 | 614155 | 614458 | E070 | 1130 |
| chr11 | 578622 | 578923 | E071 | -34102 |
| chr11 | 579058 | 579102 | E071 | -33923 |
| chr11 | 579186 | 579249 | E071 | -33776 |
| chr11 | 579295 | 579830 | E071 | -33195 |
| chr11 | 579896 | 579951 | E071 | -33074 |
| chr11 | 580027 | 580591 | E071 | -32434 |
| chr11 | 585460 | 585564 | E071 | -27461 |
| chr11 | 587478 | 588405 | E071 | -24620 |
| chr11 | 614155 | 614458 | E071 | 1130 |
| chr11 | 628527 | 628893 | E071 | 15502 |
| chr11 | 629062 | 629215 | E071 | 16037 |
| chr11 | 632579 | 632671 | E071 | 19554 |
| chr11 | 632853 | 633265 | E071 | 19828 |
| chr11 | 658284 | 658350 | E071 | 45259 |
| chr11 | 658432 | 658924 | E071 | 45407 |
| chr11 | 573158 | 573208 | E072 | -39817 |
| chr11 | 573276 | 573473 | E072 | -39552 |
| chr11 | 573527 | 573571 | E072 | -39454 |
| chr11 | 573577 | 573647 | E072 | -39378 |
| chr11 | 573746 | 573796 | E072 | -39229 |
| chr11 | 578622 | 578923 | E072 | -34102 |
| chr11 | 579058 | 579102 | E072 | -33923 |
| chr11 | 579186 | 579249 | E072 | -33776 |
| chr11 | 579295 | 579830 | E072 | -33195 |
| chr11 | 579896 | 579951 | E072 | -33074 |
| chr11 | 606022 | 606109 | E072 | -6916 |
| chr11 | 606130 | 606170 | E072 | -6855 |
| chr11 | 606272 | 606606 | E072 | -6419 |
| chr11 | 606695 | 606803 | E072 | -6222 |
| chr11 | 614155 | 614458 | E072 | 1130 |
| chr11 | 632510 | 632560 | E072 | 19485 |
| chr11 | 632579 | 632671 | E072 | 19554 |
| chr11 | 632853 | 633265 | E072 | 19828 |
| chr11 | 658284 | 658350 | E072 | 45259 |
| chr11 | 658432 | 658924 | E072 | 45407 |
| chr11 | 578622 | 578923 | E073 | -34102 |
| chr11 | 579058 | 579102 | E073 | -33923 |
| chr11 | 579186 | 579249 | E073 | -33776 |
| chr11 | 579295 | 579830 | E073 | -33195 |
| chr11 | 579896 | 579951 | E073 | -33074 |
| chr11 | 658432 | 658924 | E073 | 45407 |
| chr11 | 573158 | 573208 | E074 | -39817 |
| chr11 | 573276 | 573473 | E074 | -39552 |
| chr11 | 573527 | 573571 | E074 | -39454 |
| chr11 | 573577 | 573647 | E074 | -39378 |
| chr11 | 578622 | 578923 | E074 | -34102 |
| chr11 | 579058 | 579102 | E074 | -33923 |
| chr11 | 579186 | 579249 | E074 | -33776 |
| chr11 | 579295 | 579830 | E074 | -33195 |
| chr11 | 579896 | 579951 | E074 | -33074 |
| chr11 | 580027 | 580591 | E074 | -32434 |
| chr11 | 614155 | 614458 | E074 | 1130 |
| chr11 | 616501 | 617304 | E074 | 3476 |
| chr11 | 658284 | 658350 | E074 | 45259 |
| chr11 | 658432 | 658924 | E074 | 45407 |
| chr11 | 569960 | 570100 | E081 | -42925 |
| chr11 | 570113 | 570215 | E081 | -42810 |
| chr11 | 578622 | 578923 | E081 | -34102 |
| chr11 | 579058 | 579102 | E081 | -33923 |
| chr11 | 579186 | 579249 | E081 | -33776 |
| chr11 | 579295 | 579830 | E081 | -33195 |
| chr11 | 579896 | 579951 | E081 | -33074 |
| chr11 | 580027 | 580591 | E081 | -32434 |
| chr11 | 614155 | 614458 | E081 | 1130 |
| chr11 | 578622 | 578923 | E082 | -34102 |
| chr11 | 579058 | 579102 | E082 | -33923 |
| chr11 | 579186 | 579249 | E082 | -33776 |
| chr11 | 579295 | 579830 | E082 | -33195 |
| chr11 | 579896 | 579951 | E082 | -33074 |
| chr11 | 614155 | 614458 | E082 | 1130 |
Promoter Annotation (GRCh37.p13)
| Chromosome | Start | End | Region | Distance(-/+:Up/Downstream) |
|---|---|---|---|---|
| chr11 | 567281 | 569927 | E067 | -43098 |
| chr11 | 574250 | 576772 | E067 | -36253 |
| chr11 | 576899 | 577749 | E067 | -35276 |
| chr11 | 614468 | 616352 | E067 | 1443 |
| chr11 | 567281 | 569927 | E068 | -43098 |
| chr11 | 574250 | 576772 | E068 | -36253 |
| chr11 | 576899 | 577749 | E068 | -35276 |
| chr11 | 614468 | 616352 | E068 | 1443 |
| chr11 | 567281 | 569927 | E069 | -43098 |
| chr11 | 574250 | 576772 | E069 | -36253 |
| chr11 | 576899 | 577749 | E069 | -35276 |
| chr11 | 567281 | 569927 | E070 | -43098 |
| chr11 | 574250 | 576772 | E070 | -36253 |
| chr11 | 576899 | 577749 | E070 | -35276 |
| chr11 | 614468 | 616352 | E070 | 1443 |
| chr11 | 567281 | 569927 | E071 | -43098 |
| chr11 | 574250 | 576772 | E071 | -36253 |
| chr11 | 576899 | 577749 | E071 | -35276 |
| chr11 | 614468 | 616352 | E071 | 1443 |
| chr11 | 567281 | 569927 | E072 | -43098 |
| chr11 | 574250 | 576772 | E072 | -36253 |
| chr11 | 576899 | 577749 | E072 | -35276 |
| chr11 | 567281 | 569927 | E073 | -43098 |
| chr11 | 574250 | 576772 | E073 | -36253 |
| chr11 | 576899 | 577749 | E073 | -35276 |
| chr11 | 614468 | 616352 | E073 | 1443 |
| chr11 | 567281 | 569927 | E074 | -43098 |
| chr11 | 574250 | 576772 | E074 | -36253 |
| chr11 | 576899 | 577749 | E074 | -35276 |
| chr11 | 614468 | 616352 | E074 | 1443 |
| chr11 | 576899 | 577749 | E081 | -35276 |
| chr11 | 567281 | 569927 | E082 | -43098 |
| chr11 | 574250 | 576772 | E082 | -36253 |
| chr11 | 576899 | 577749 | E082 | -35276 |
| chr11 | 614468 | 616352 | E082 | 1443 |
| chr11 | 636787 | 637366 | E082 | 23762 |
| chr11 | 639624 | 639981 | E082 | 26599 |
| chr11 | 640123 | 640340 | E082 | 27098 |